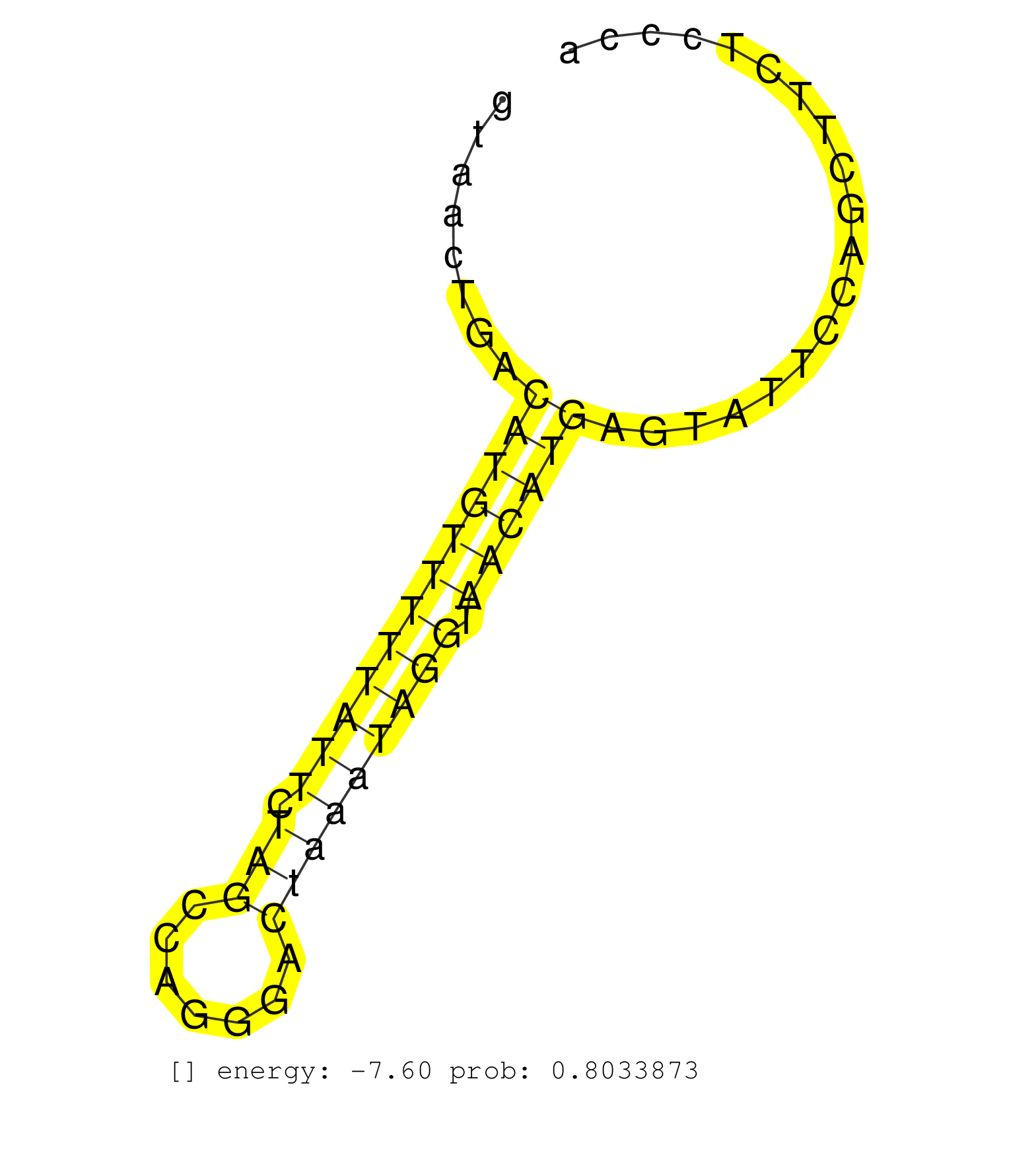
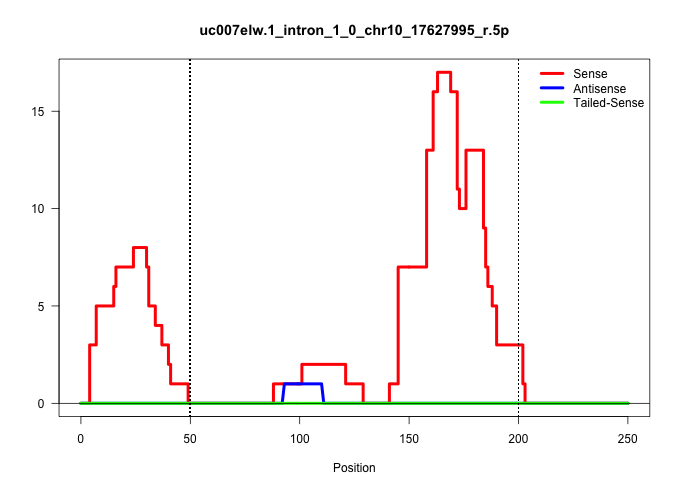
| Gene: Heca | ID: uc007elw.1_intron_1_0_chr10_17627995_r.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(22) TESTES |
| CCCCTCAAGACACGATGAGATTGAATATGATGTTCCTTGTCACCTTCAAGGTATAGGTGTTAATTCACCTTGCCTGGTTTCTATTTGTTAAAACCCAAAAAAGCAAACAAGATGGAACTACATTTTTACTTTATTTAATTGTAACTGACATGTTTTTATTCTAGCCAGGGACTAAATAGGTAACATGAGTATTCCAGCTTCTCCCAGTGGCTGGTGGTTTTTGTACTTGGATCTCACTGTACTGGGTACA ....................................................................................................................................................((((((((((((.(((.......))))))))).))))))............................................................... ............................................................................................................................................141..............................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TGACATGTTTTTATTCTAGCCAGGGAC.............................................................................. | 27 | 1 | 5.00 | 5.00 | - | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TTCTAGCCAGGGACTAAATAGGTAAC.................................................................. | 26 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCAAGACACGATGAGATTGAATATGAT........................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAGGTAACATGAGTATTCCAGCTTCT................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................TTCTAGCCAGGGACTAAATAGGTAACA................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TGAGATTGAATATGATGTTCCTTGTC................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCCAGGGACTAAATAGGTAACATGAGT............................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGACATGTTTTTATTCTAGCCAGGGACT............................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TAGCCAGGGACTAAATAGGTAACA................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......AGACACGATGAGATTGAATATGATGTT........................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAGGTAACATGAGTATTCCAGCTTCTC............................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................ATATGATGTTCCTTGTCACCTTCAA......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................AGCAAACAAGATGGAACTACATTTTTAC......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................TAGCCAGGGACTAAATAGGTAACATGA.............................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................TAGCCAGGGACTAAATAGGTAACATGAGT............................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TTCTAGCCAGGGACTAAATAGGTAACAT................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......AGACACGATGAGATTGAATATGATGTTCCT..................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................TAAAACCCAAAAAAGCAAACAAGATGGAACTAC................................................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TAACTGACATGTTTTTATTCTAGCCAGG................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................ACTTGGATCTCACTGTACTGGGTACA | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................GAGATTGAATATGATGTTCCTTGT.................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....TCAAGACACGATGAGATTGAATATGA............................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCCTCAAGACACGATGAGATTGAATATGATGTTCCTTGTCACCTTCAAGGTATAGGTGTTAATTCACCTTGCCTGGTTTCTATTTGTTAAAACCCAAAAAAGCAAACAAGATGGAACTACATTTTTACTTTATTTAATTGTAACTGACATGTTTTTATTCTAGCCAGGGACTAAATAGGTAACATGAGTATTCCAGCTTCTCCCAGTGGCTGGTGGTTTTTGTACTTGGATCTCACTGTACTGGGTACA ....................................................................................................................................................((((((((((((.(((.......))))))))).))))))............................................................... ............................................................................................................................................141..............................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR037900(GSM510436) testes_rep1. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................CCCAAAAAAGCAAACAgc............................................................................................................................................. | 18 | gc | 4.00 | 0.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CCCAAAAAAGCAAAgaat............................................................................................................................................... | 18 | gaat | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................CCCAAAAAAGCAAACAAGgc........................................................................................................................................... | 20 | gc | 2.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................CCCAAAAAAGCAAACAAGgcc........................................................................................................................................... | 21 | gcc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CCCAAAAAAGCAAAgcat............................................................................................................................................... | 18 | gcat | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................CCCAAAAAAGCAAACAgcc............................................................................................................................................. | 19 | gcc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................CCCAAAAAAGCAAACAAgccc............................................................................................................................................ | 21 | gccc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................CCCAAAAAAGCAAACAAG........................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |