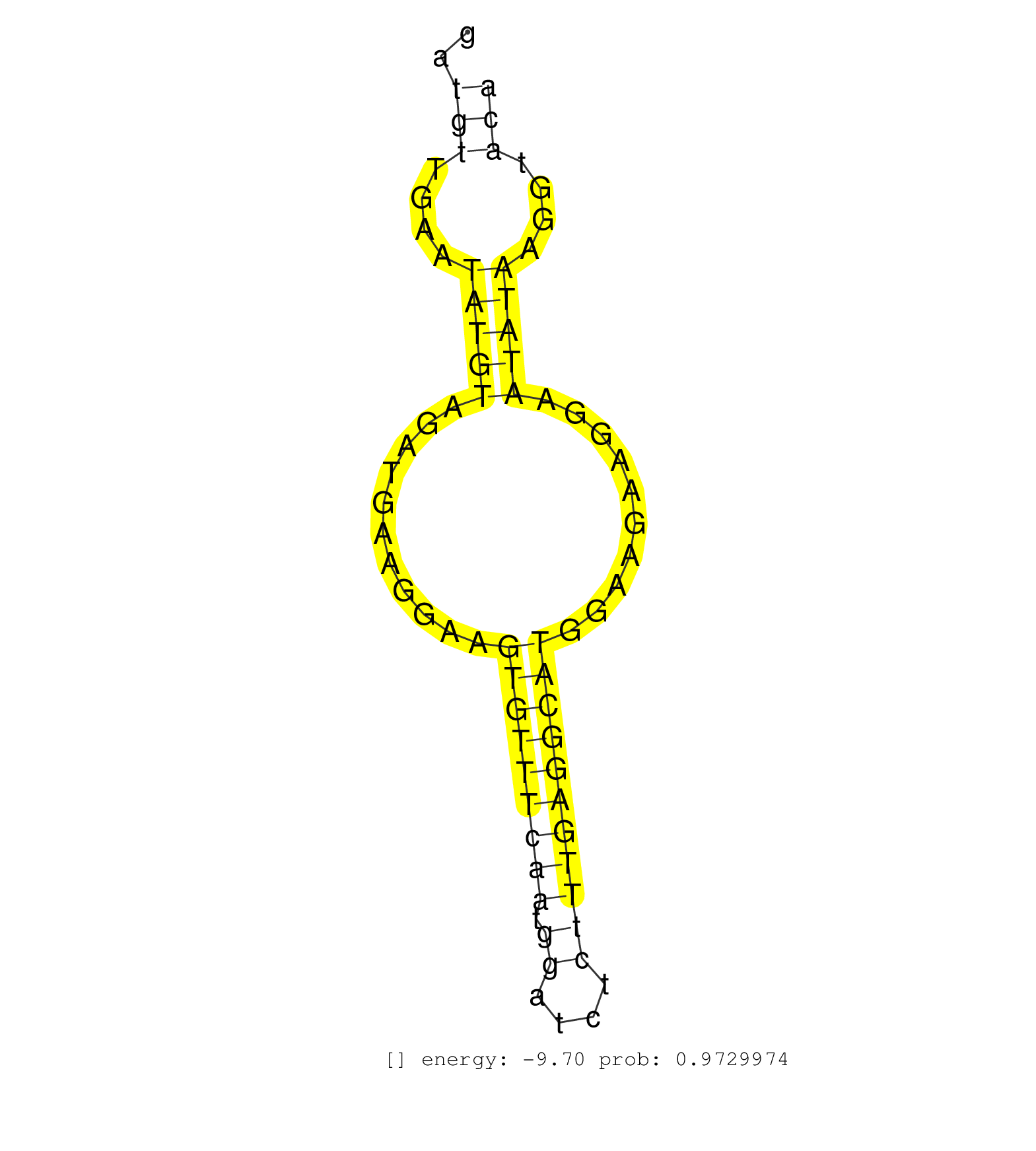

| Gene: Shprh | ID: uc007ejr.1_intron_11_0_chr10_10907835_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(13) TESTES |
| AAGGACTTGTAGATGATAGAGTACCTACCACTACCAGAGGTCTGTGGGCCGTAAGTGAGACGGAGCGATCTATGAAAGCAATATTATCATTTGCAAGATCACATAGGTTTGATGTTGAATATGTAGATGAAGGAAGTGTTTCAATGGATCTCTTTGAGGCATGGAAGAAGGAATATAAGGTACACTATTTTTAAACTTCTGAAATTAAAAACCTTTTTGCATTAACTTGAAAATCTGCTTTGGAGACTGT ................................................................................................................(((....(((((...........(((((((((.((....)))))))))))..........)))))....))).................................................................. ..............................................................................................................111......................................................................184................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................TTGAGGCATGGAAGAAGGAATATAAGt...................................................................... | 27 | t | 9.00 | 0.00 | 4.00 | 3.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................TGAATATGTAGATGAAGGAAGTGTTT............................................................................................................. | 26 | 1 | 5.00 | 5.00 | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................TTGAGGCATGGAAGAAGGAATATAAGtt..................................................................... | 28 | tt | 4.00 | 0.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAATATGTAGATGAAGGAAGTGTTTC............................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TGAATATGTAGATGAAGGAAGTGTT.............................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 |
| ........................................................................................................................................................TTTGAGGCATGGAAGAAGGAATATAAGt...................................................................... | 28 | t | 3.00 | 0.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TTTGAGGCATGGAAGAAGGAATATAAGtt..................................................................... | 29 | tt | 3.00 | 0.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................CGTAAGTGAGACGGAGCGATCTATGA............................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................CTTTGAGGCATGGAAGAAGGAATATAAGt...................................................................... | 29 | t | 2.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TGATGTTGAATATGTAGATGAAGGAA................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGAGGCATGGAAGAAGGAATATAAGtt..................................................................... | 27 | tt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................TACCAGAGGTCTGTGGGCact...................................................................................................................................................................................................... | 21 | act | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................GAATATGTAGATGAAGGAAGTGTTT............................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TATCATTTGCAAGATCACATAGGTTTGA.......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................ATATGTAGATGAAGGAAGTG................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................TTTGAGGCATGGAAGAAGGAATATAA........................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........AGATGATAGAGTACCTACCAC........................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................ATGTTGAATATGTAGATG......................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CTTTGAGGCATGGAAGAAGGAATATA......................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TTGAGGCATGGAAGAAGG............................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................TCTTTGAGGCATGGAAGAAGGAATAT.......................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TTGAGGCATGGAAGAAGGAATATAAGtta.................................................................... | 29 | tta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TAGGTTTGATGTTGAATATGTAGATGAAG...................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................TGAGACGGAGCGATCTATGAAAGCAATA....................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................GTTTGATGTTGAATATGTAGAT.......................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................GTTTGATGTTGAATATGTAGATGAAGGA.................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGTGAGACGGAGCGATCTATGAAAGC........................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGGACTTGTAGATGATAGAGTACCTACCACTACCAGAGGTCTGTGGGCCGTAAGTGAGACGGAGCGATCTATGAAAGCAATATTATCATTTGCAAGATCACATAGGTTTGATGTTGAATATGTAGATGAAGGAAGTGTTTCAATGGATCTCTTTGAGGCATGGAAGAAGGAATATAAGGTACACTATTTTTAAACTTCTGAAATTAAAAACCTTTTTGCATTAACTTGAAAATCTGCTTTGGAGACTGT ................................................................................................................(((....(((((...........(((((((((.((....)))))))))))..........)))))....))).................................................................. ..............................................................................................................111......................................................................184................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR042485(GSM539877) mouse testicular tissue [09-002]. (testes) |
|---|