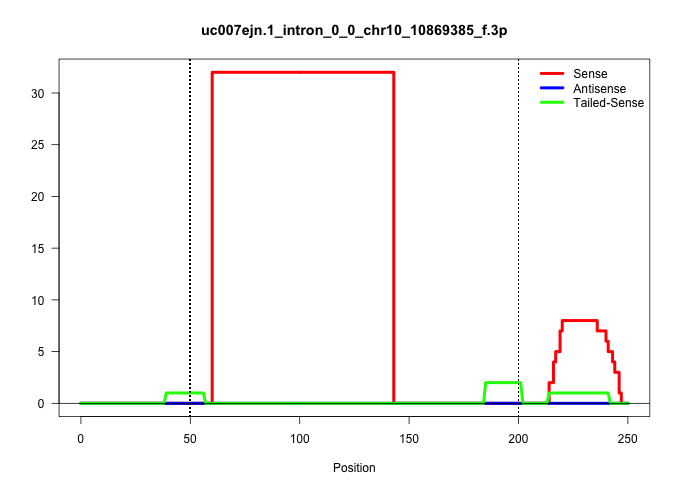
| Gene: Shprh | ID: uc007ejn.1_intron_0_0_chr10_10869385_f.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(2) PIWI.mut |
(10) TESTES |
| TTGTTTGGAACTATTTCCTCATAAAAGATATATCACCTCAGTGTTGGTAACCTTCACAGCAGATGTACATTGTTAATTTTCCTGGAGTCTAGATTTGCAGTTAGCCAGAGGCTTTAACCAGACTTGACTGTATGGACTCTATTCTCTTAGTTTTCCTGTCTTTATCCTGCCTCACACCAACTATTCCATTTGTCATTTAGAATTCACATTTACTTATCAAGAAAGTGGAAGCGTGAAAATGAGCAGCCGA ..........................................................(((((((......(((((((...((((...))))......)))))))..(((((.......((((..(((((...(((......)))..))))).....)))).......))))).............)))))))......................................................... .......................................................56....................................................................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................AGATGTACATTGTTAATTTTCCTGGAGTCTAGATTTGCAGTTAGCCAGAGGC.......................................................................................................................................... | 52 | 1 | 32.00 | 32.00 | 32.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTGGAAGCGTGAAAATGAGCAG.... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................CCATTTGTCATTTttgg................................................ | 17 | ttgg | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - |
| .......................................AGTGTTGGTAACCTctgg................................................................................................................................................................................................. | 18 | ctgg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................................................................................................TCAAGAAAGTGGAAGCGTGAAAATGAGC...... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................................TATCAAGAAAGTGGAAGCGTGAAAATGt........ | 28 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................................TATCAAGAAAGTGGAAGCGTGAAAAT.......... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GAAAGTGGAAGCGTGAAAATGAGCAGC... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................CAAGAAAGTGGAAGCGTGA.............. | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TCAAGAAAGTGGAAGCGTGAAAATGAG....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ......................................................................................................................................................................................................................TATCAAGAAAGTGGAAGCGTGAAAATG......... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| TTGTTTGGAACTATTTCCTCATAAAAGATATATCACCTCAGTGTTGGTAACCTTCACAGCAGATGTACATTGTTAATTTTCCTGGAGTCTAGATTTGCAGTTAGCCAGAGGCTTTAACCAGACTTGACTGTATGGACTCTATTCTCTTAGTTTTCCTGTCTTTATCCTGCCTCACACCAACTATTCCATTTGTCATTTAGAATTCACATTTACTTATCAAGAAAGTGGAAGCGTGAAAATGAGCAGCCGA ..........................................................(((((((......(((((((...((((...))))......)))))))..(((((.......((((..(((((...(((......)))..))))).....)))).......))))).............)))))))......................................................... .......................................................56....................................................................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................GTAACCTTCACAGCAact............................................................................................................................................................................................. | 18 | act | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................AGCAGATGTACATTGTTAATtgt............................................................................................................................................................................. | 23 | tgt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - |