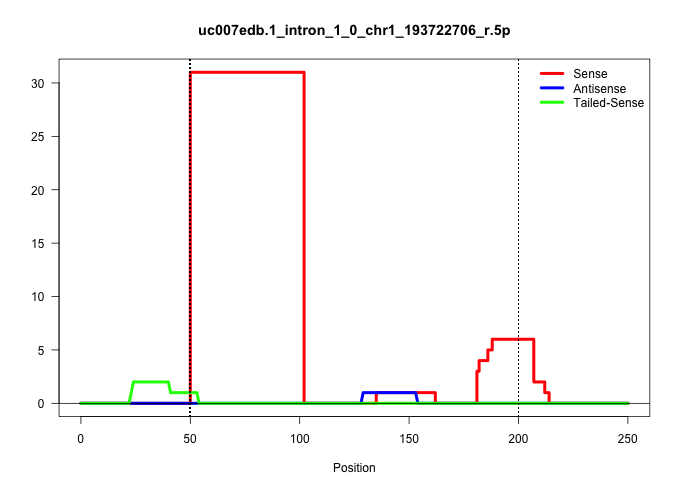
| Gene: AK137370 | ID: uc007edb.1_intron_1_0_chr1_193722706_r.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(9) TESTES |
| CTACAGAGTCTGCTTCTCCCTGCTTCCTGTTCCTCTACACCGCTGGCTCTGTGAGTCTTTCGTTAATTTGTAATTACTATAAGCACTAAGGGGAGGTGGCCTACTTGCTAGTGACACCTCTCTTTAACAGGTCACTCCAAGGTCTGATTCTCTAGTGACGCATTCCAAAATTGGGTCCTCTTTACATATAGTTCTGTGGGGTGAATTAGTGCCATGTTCTGCCTGCTGCTTTGGTCAGTGTAACCACTAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTCTTTCGTTAATTTGTAATTACTATAAGCACTAAGGGGAGGTGGCCT.................................................................................................................................................... | 52 | 1 | 31.00 | 31.00 | 31.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TTACATATAGTTCTGTGGGGTGAATT........................................... | 26 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - |
| ........................TCCTGTTCCTCTACACCGCTGGCTCTatgt.................................................................................................................................................................................................... | 30 | atgt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................TCCAAGGTCTGATTCTCTAGTGACGCA........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| .......................TTCCTGTTCCTCTAgggt................................................................................................................................................................................................................. | 18 | gggt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................................................TACATATAGTTCTGTGGGGTGAATT........................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................................TAGTTCTGTGGGGTGAATTAGTGCCA.................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................TATAGTTCTGTGGGGTGAATTAGTGC...................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| CTACAGAGTCTGCTTCTCCCTGCTTCCTGTTCCTCTACACCGCTGGCTCTGTGAGTCTTTCGTTAATTTGTAATTACTATAAGCACTAAGGGGAGGTGGCCTACTTGCTAGTGACACCTCTCTTTAACAGGTCACTCCAAGGTCTGATTCTCTAGTGACGCATTCCAAAATTGGGTCCTCTTTACATATAGTTCTGTGGGGTGAATTAGTGCCATGTTCTGCCTGCTGCTTTGGTCAGTGTAACCACTAC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................GGTCACTCCAAGGTCTGATTCTCTA................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................TCTGATTCTCTAGTagct.............................................................................................. | 18 | agct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |