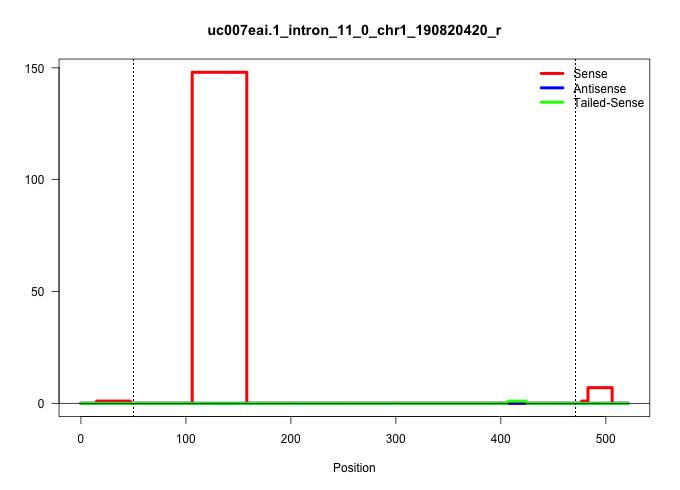
| Gene: Kctd3 | ID: uc007eai.1_intron_11_0_chr1_190820420_r | SPECIES: mm9 |
 |
|
(1) PIWI.ip |
(1) PIWI.mut |
(8) TESTES |
| GAGGTCATCCTGCGGCAGCGTCCTGTTCCACGGCTACTTGCCCCCGCCAGGTACTGCAGTCTTCAGAAAATTCCTCTTGGCACTGAGCATAAGCATAAAAATAAGAAAACTGTTGGTTATTTGAGTTTGGATAAATCTGGACCTCATCAGACCGTGCCCTTGCATGAGAGTGAAGCTGCACGGTCTGTAGAGCAGGGCTGGCGGCTTCTCTGTCAGGGCAGGGAGGAGCTCCGTGGCCTCACTCAGCTGCCATGTATACCACTGGTCCTTGTGTGTGTGGGGGTGGGTGGGTGGGGGATGGCACGGGACCTGAGCCTGGGGCTCTGCTTAGGTCACCGTCCCCTCAGTTACTTACTAGAGTTGGGCCTTTGTGCACCATTGTGAGAGAGAGGTGTGGTTTTTGAAAAGATCCTGTTTGAAAGTATGAGGAAATTCAAGCATAACAGATAATATGTGTATTCTCCTTTGTAGGCATTCCCAGTCGTAAAGTAACTAACACAGCTAGGTCCTCTGTGGATGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................AAACTGTTGGTTATTTGAGTTTGGATAAATCTGGACCTCATCAGACCGTGCC........................................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 148.00 | 148.00 | 148.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................GTAAAGTAACTAACACAGCTAGG............... | 23 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................GATCCTGTTTGAAAGagt................................................................................................ | 18 | agt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CCAGTCGTAAAGTAACTAACACAGCTAGG............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ...............CAGCGTCCTGTTCCACGGCTACTTGCCCCCGC.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| GAGGTCATCCTGCGGCAGCGTCCTGTTCCACGGCTACTTGCCCCCGCCAGGTACTGCAGTCTTCAGAAAATTCCTCTTGGCACTGAGCATAAGCATAAAAATAAGAAAACTGTTGGTTATTTGAGTTTGGATAAATCTGGACCTCATCAGACCGTGCCCTTGCATGAGAGTGAAGCTGCACGGTCTGTAGAGCAGGGCTGGCGGCTTCTCTGTCAGGGCAGGGAGGAGCTCCGTGGCCTCACTCAGCTGCCATGTATACCACTGGTCCTTGTGTGTGTGGGGGTGGGTGGGTGGGGGATGGCACGGGACCTGAGCCTGGGGCTCTGCTTAGGTCACCGTCCCCTCAGTTACTTACTAGAGTTGGGCCTTTGTGCACCATTGTGAGAGAGAGGTGTGGTTTTTGAAAAGATCCTGTTTGAAAGTATGAGGAAATTCAAGCATAACAGATAATATGTGTATTCTCCTTTGTAGGCATTCCCAGTCGTAAAGTAACTAACACAGCTAGGTCCTCTGTGGATGCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR037900(GSM510436) testes_rep1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTCCCAGTCGTcggc.................................... | 15 | cggc | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - |
| .............................................GGTACTGCAGTCTTagat.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 18 | agat | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................ACGGTCTGTAGAGCcgcc........................................................................................................................................................................................................................................................................................................................................ | 18 | cgcc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - |