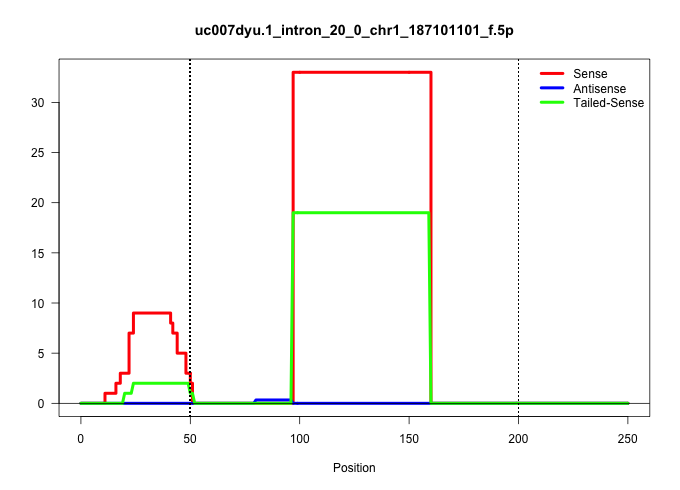
| Gene: Rab3gap2 | ID: uc007dyu.1_intron_20_0_chr1_187101101_f.5p | SPECIES: mm9 |
 |
 |
|
(2) PIWI.ip |
(12) TESTES |
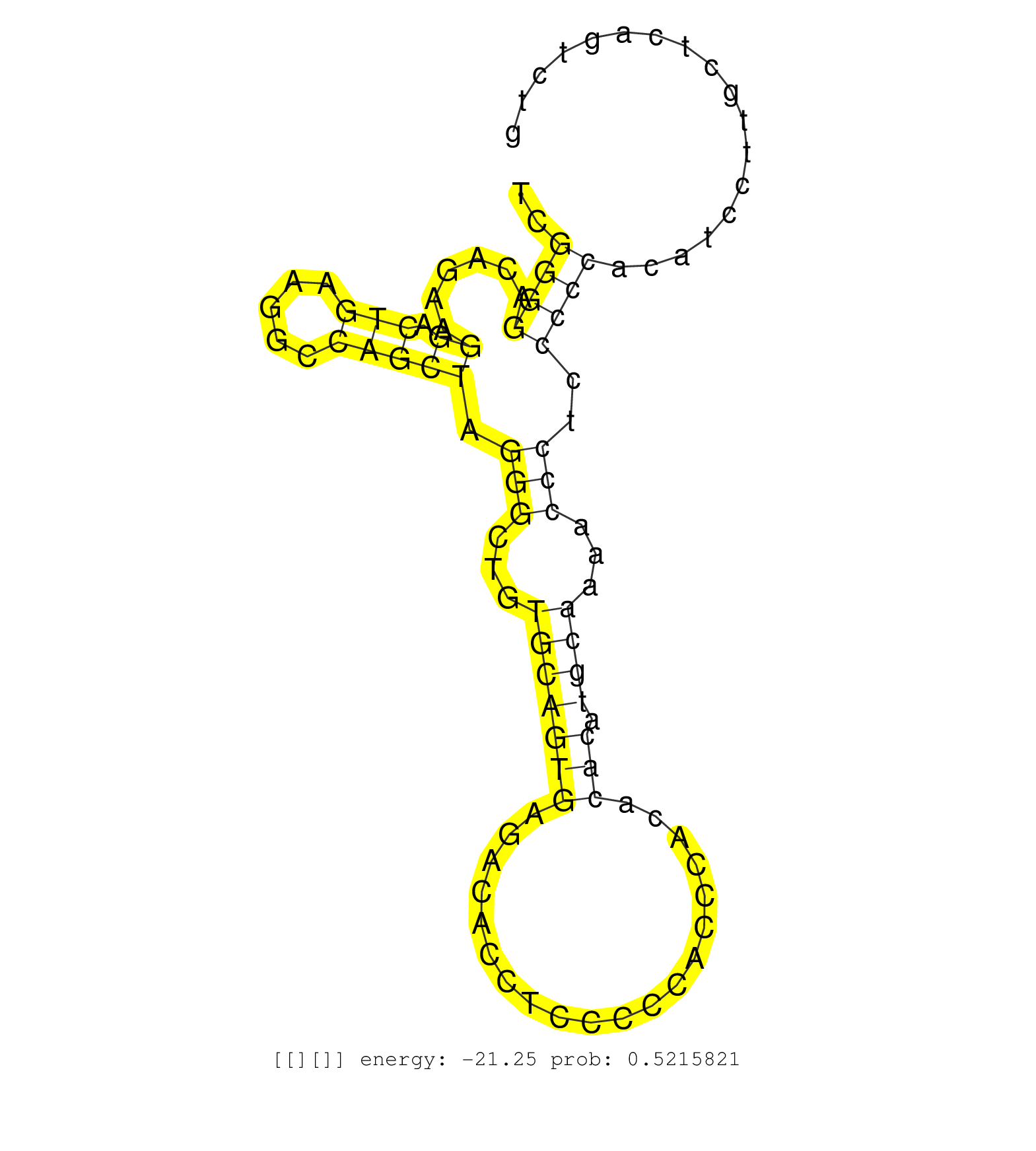
| TGACATCCTTCCTCGGCTCCTGCCTGGAACTTCTTCAGACGTCATTGGAGGTGAACACTAGCCATGTGGTGACGCGTTGCCTGTACTCTCCTCCTGGGCATCGGGGACAGAAGGACTGAAGGCCAGCTAGGGCTGTGCAGTGAGACACCTCCCCCACCCACACACATGCAAAACCCTCCCCCACATCCTTGCTCAGTCTGAGTGTCTCTTTAGACATTAGATGTCCTGAAAACATTCAGTTTAAGAGATG ......................................................................................................((((......((.(((.....))))).(((...(((((((....................))).))))...)))..)))).................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................GCATCGGGGACAGAAGGACTGAAGGCCAGCTAGGGCTGTGCAGTGAGACACC..................................................................................................... | 52 | 1 | 53.00 | 53.00 | 53.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................CCTGGAACTTCTTCAGACGTCATTGG.......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - |
| ......................CCTGGAACTTCTTCAGACGTCATTGGAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........CTCGGCTCCTGCCTGGAACTTCTTCAGACG................................................................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................TGGAACTTCTTCAGACGTCA.............................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................TGGAACTTCTTCAGACGTCATTGGAGGc...................................................................................................................................................................................................... | 28 | c | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................TGGAACTTCTTCAGACGTCATTGGAGG....................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................CCTGCCTGGAACTTCTTCAGACGT................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................GCATCGGGGACAGAAGGACTGAAG................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................CCTGGAACTTCTTCAGACGTCATTGGAGG....................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................TGCCTGGAACTTCTTCAGACGTCATTGttt........................................................................................................................................................................................................ | 30 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................CTCCTGCCTGGAACTTCTTCAGACGTCA.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TGACATCCTTCCTCGGCTCCTGCCTGGAACTTCTTCAGACGTCATTGGAGGTGAACACTAGCCATGTGGTGACGCGTTGCCTGTACTCTCCTCCTGGGCATCGGGGACAGAAGGACTGAAGGCCAGCTAGGGCTGTGCAGTGAGACACCTCCCCCACCCACACACATGCAAAACCCTCCCCCACATCCTTGCTCAGTCTGAGTGTCTCTTTAGACATTAGATGTCCTGAAAACATTCAGTTTAAGAGATG ......................................................................................................((((......((.(((.....))))).(((...(((((((....................))).))))...)))..)))).................................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................CTGTACTCTCCTCCTGG......................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | 0.33 |