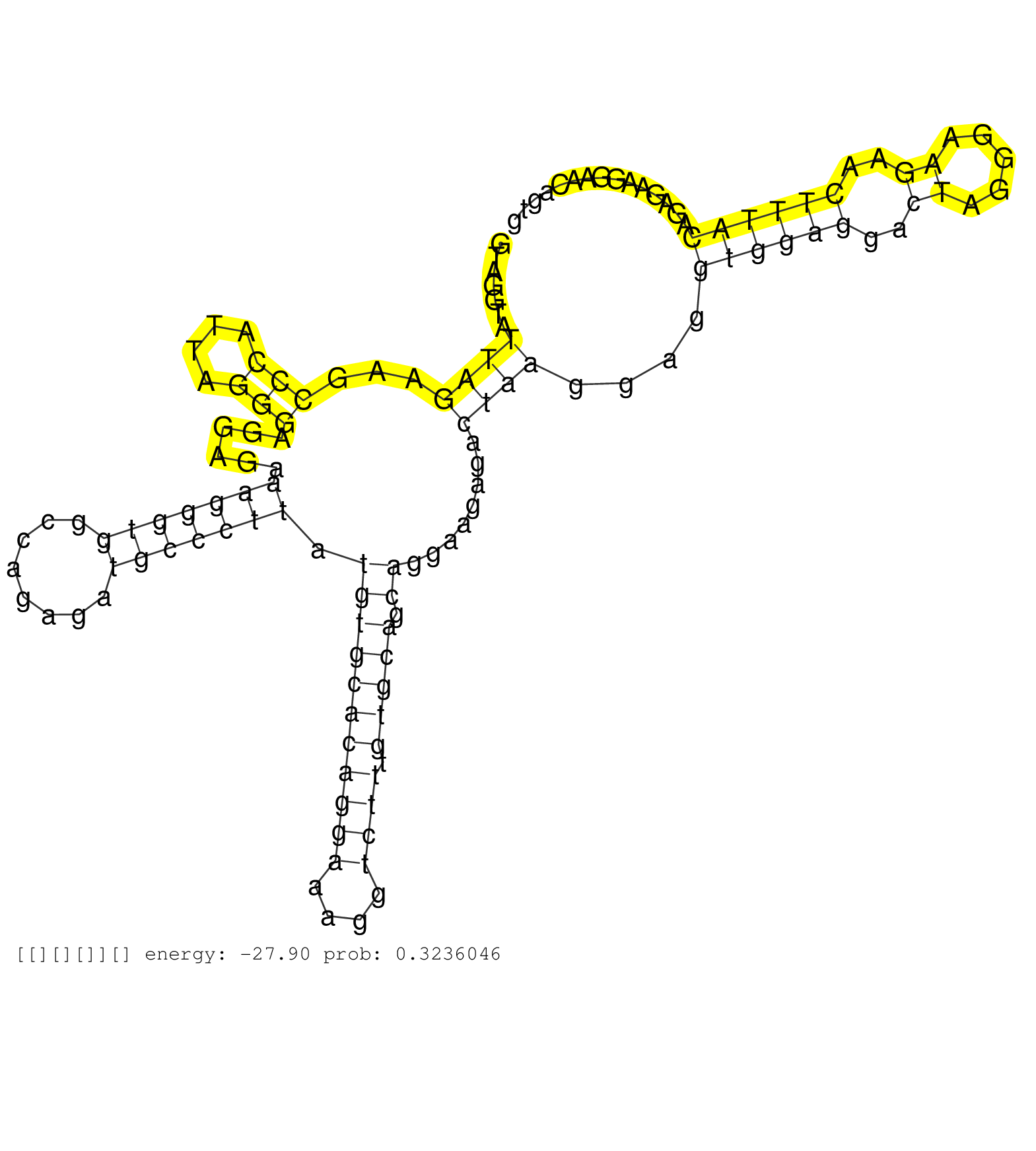

| Gene: Lbr | ID: uc007dxo.1_intron_8_0_chr1_183759040_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(20) TESTES |
| TCAAGTGACCACTCCACAGAGGAAGGACTTGGAGTTTGGAGGAGTACCTGGTAGGTATTAGAAGCCCATTAGGGAGGAGAAAGGGTGGCCAGAGATGCCCTTATGTGCACAGGAAAGGTCTTTGTGCAGCAGGAAGAGACTAAGGAGGTGGAGGACTAGGGAAGAACTTTACAGAGAAGGAACAGTGGTATGAAGGCCTGAGCTGGGAGCTACGGCAGTGTGTGTACAGGGCACAACTGGGGTGGTACAT .........................................................((((...(((....)))......(((((((........))))))).(((((((((((....)))).))))).))........))))....((((((..((.....))..)))))).............................................................................. ..................................................51......................................................................................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................TAGGGAAGAACTTTACAGAGAAGGAAC................................................................... | 27 | 1 | 15.00 | 15.00 | 1.00 | 5.00 | 4.00 | 1.00 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TACAGAGAAGGAACAGTGGTATGAAGGC..................................................... | 28 | 1 | 14.00 | 14.00 | 3.00 | 1.00 | 6.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATTAGAAGCCCATTAGGGAGGAG........................................................................................................................................................................... | 29 | 1 | 12.00 | 12.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATTAGAAGCCCATTAGGGAGGAGA.......................................................................................................................................................................... | 30 | 1 | 8.00 | 8.00 | 3.00 | 2.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TGAAGGCCTGAGCTGGGAGCTACGG................................... | 25 | 1 | 6.00 | 6.00 | 2.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAGGTATTAGAAGCCCATTAGGGAGG............................................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................GGAAGGACTTGGAGTTTGGAG................................................................................................................................................................................................................. | 21 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGGAAGAACTTTACAGAGAAGGAACAGT................................................................ | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TACAGAGAAGGAACAGTGGTATGAAGG...................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AAGAACTTTACAGAGAAGGAACAGTGGT............................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TGAAGGCCTGAGCTGGGAGCTACGGC.................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AAGGACTTGGAGTTTGGAGGAGTACC.......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TAGGGAAGAACTTTACAGAGAAGGAA.................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGTATTAGAAGCCCATTAGGGAGGA............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GGGAAGAACTTTACAGAGAAGGAACAG................................................................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GTTTGGAGGAGTACCTG........................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TAGGGAAGAACTTTACAGAGAAGGAACAG................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................ACTAGGGAAGAACTTTACAGAGAAGGA..................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AGAAGGAACAGTGGTATGAAGGCCTGA................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TTAGAAGCCCATTAGGGAGGAGAAAGGG..................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................TGCAGCAGGAAGAGACTAAGGAGGTGG................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................AGGAAGGACTTGGAGTTTGGAGG................................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TTGTGCAGCAGGAAGAGACTAAGGAG....................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GAAGGAACAGTGGTATGAAGGCCTGA................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TAAGGAGGTGGAGGACTAGGGAAG...................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................TTTGTGCAGCAGGAAGAGACTAAGGAG....................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TACAGAGAAGGAACAGTGGTATGAAGGCC.................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................AGGGAAGAACTTTACAGAGAAGGAACAGT................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................GACTAGGGAAGAACTTTACAGAGA......................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AAGAACTTTACAGAGAAGGAACAGTGG.............................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GGAAGAACTTTACAGAGAAGGAACAGT................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TACAGAGAAGGAACAGTGGTATGAAG....................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................GCAGGAAGAGACTAAGGAGGTG.................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................AGGAACAGTGGTATGAAGGCCTGAGC............................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................TGGAGTTTGGAGGAGTACCTGGT...................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................AGAACTTTACAGAGAAGGAACAGTGG.............................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................TATGAAGGCCTGAGCTGGGAGCTACGG................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................TGAAGGCCTGAGCTGGGAGCTACGGta................................. | 27 | ta | 1.00 | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................CAGAGGAAGGACTTGGAGTTTGGAGG................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................GTGGTATGAAGGCCTGAGCTGGGAG......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CCTGAGCTGGGAGCTACGGCAGggt............................. | 25 | ggt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TAGGGAAGAACTTTACAGAGAAGGAACA.................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................AGGAAGGACTTGGAGTTTGGAG................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CTAGGGAAGAACTTTACAGAGAAGGAAC................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GACTTGGAGTTTGGAG................................................................................................................................................................................................................. | 16 | 5 | 0.20 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCAAGTGACCACTCCACAGAGGAAGGACTTGGAGTTTGGAGGAGTACCTGGTAGGTATTAGAAGCCCATTAGGGAGGAGAAAGGGTGGCCAGAGATGCCCTTATGTGCACAGGAAAGGTCTTTGTGCAGCAGGAAGAGACTAAGGAGGTGGAGGACTAGGGAAGAACTTTACAGAGAAGGAACAGTGGTATGAAGGCCTGAGCTGGGAGCTACGGCAGTGTGTGTACAGGGCACAACTGGGGTGGTACAT .........................................................((((...(((....)))......(((((((........))))))).(((((((((((....)))).))))).))........))))....((((((..((.....))..)))))).............................................................................. ..................................................51......................................................................................................................................187............................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................GGTCTTTGTGCAGCAcggt....................................................................................................................... | 19 | cggt | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ................................................................................................................GGTCTTTGTGCAGCgggt........................................................................................................................ | 18 | gggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................GGTCTTTGTGCAGCAgggt....................................................................................................................... | 19 | gggt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................................................................................TGTGTACAGGGCagca.................. | 16 | agca | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GGTCTTTGTGCAGCAcag....................................................................................................................... | 18 | cag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................GGTCTTTGTGCAGCAcgt....................................................................................................................... | 18 | cgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |