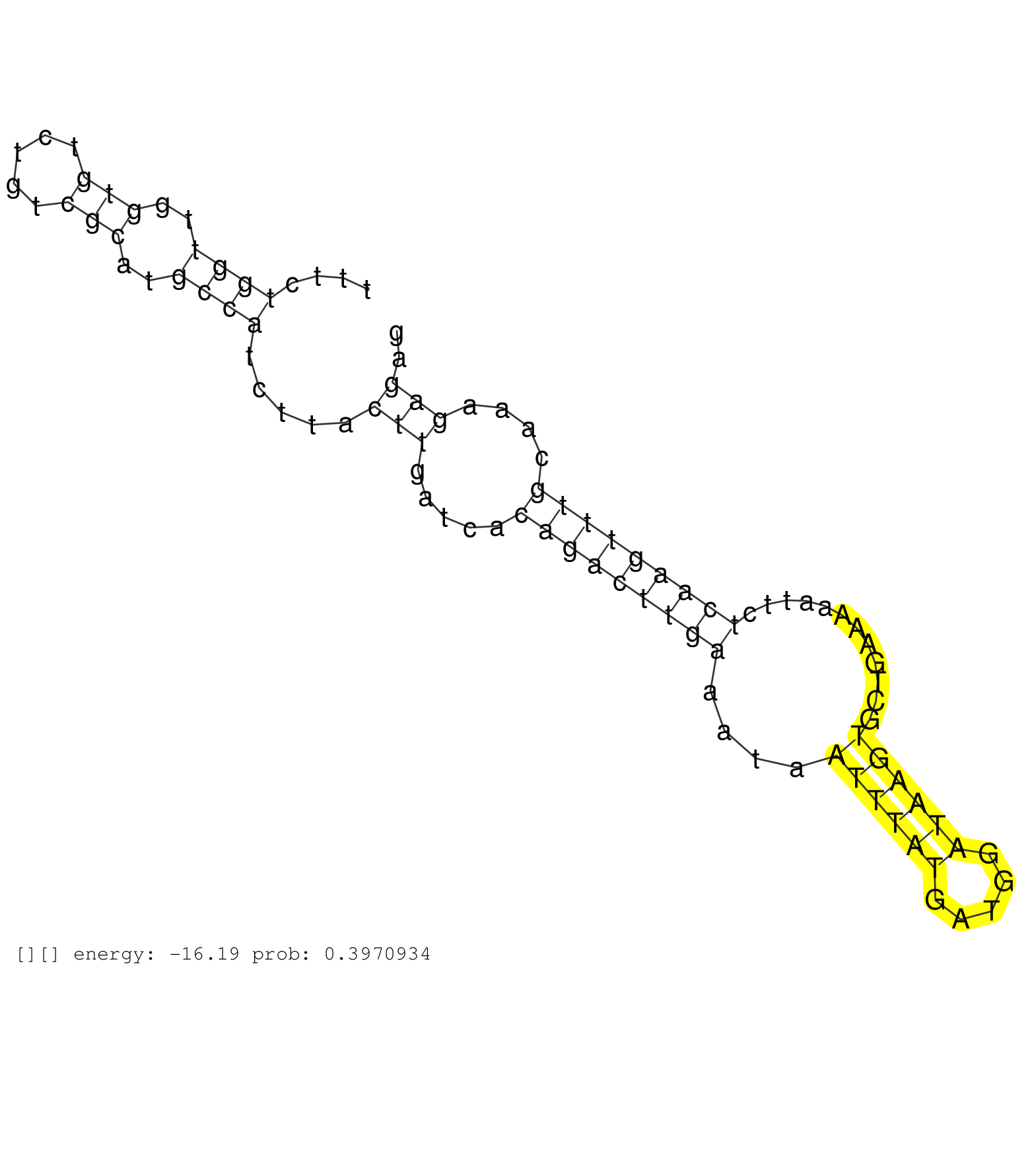
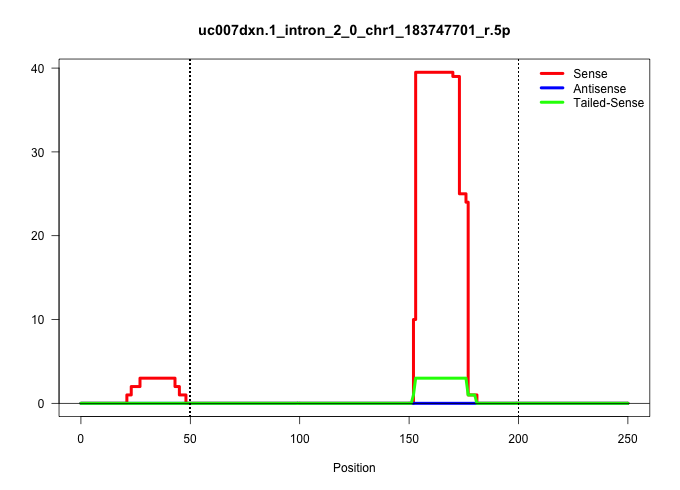
| Gene: Lbr | ID: uc007dxn.1_intron_2_0_chr1_183747701_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(12) TESTES |
| TCCCCATGACTTGTCCTGGCCATTGGCTTCTGTCATCATTGCTCTGAAATGTGAGTGCTAGTCTTAGCCTCTGTTCAGTCCAAGGCACTGTGTTTACTGTTTTCTGGTTGGTGTCTGTCGCATGCCATCTTACTTGATCACAGACTTGAAATAATTTATGATGGATAAGTGCTGAAAAATTCTCAAGTTTGCAAAGAGAGGGAATACATGCTAGCTCTCCACTAGTTCTTACAACAGACATCTTTTTTTT ........................................................................................................((((..(((.....)))..)))).....(((.....(((((((((....((((((.....))))))............)))))))))....))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................ATTTATGATGGATAAGTGCTGAAA......................................................................... | 24 | 1 | 17.00 | 17.00 | 3.00 | - | 3.00 | 4.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | 1.00 | - |
| .........................................................................................................................................................ATTTATGATGGATAAGTGCT............................................................................. | 20 | 1 | 10.00 | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................CCAAGGCACTGTGTTTACT........................................................................................................................................................ | 19 | 1 | 8.00 | 8.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................AATTTATGATGGATAAGTGCTGAAA......................................................................... | 25 | 1 | 6.00 | 6.00 | - | - | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................................................AATTTATGATGGATAAGTGCT............................................................................. | 21 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................ATTTATGATGGATAAGTGCTGcaa......................................................................... | 24 | caa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................ATTTATGATGGATAAGTGCTGAta......................................................................... | 24 | ta | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................ATTTATGATGGATAAGTGCTGAAAAATT..................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................TTCTGTCATCATTGCTCTGAA.......................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GTTTACTGTTTTCTGGg.............................................................................................................................................. | 17 | g | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................ATTTATGATGGATAAGTGCTGAA.......................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................TGGCTTCTGTCATCATTGCTCT............................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................ATTGGCTTCTGTCATCATTGCT............................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................AATTTATGATGGATAAGTGCTGAAAAATa..................................................................... | 29 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................ATTTATGATGGATAAGT................................................................................ | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| TCCCCATGACTTGTCCTGGCCATTGGCTTCTGTCATCATTGCTCTGAAATGTGAGTGCTAGTCTTAGCCTCTGTTCAGTCCAAGGCACTGTGTTTACTGTTTTCTGGTTGGTGTCTGTCGCATGCCATCTTACTTGATCACAGACTTGAAATAATTTATGATGGATAAGTGCTGAAAAATTCTCAAGTTTGCAAAGAGAGGGAATACATGCTAGCTCTCCACTAGTTCTTACAACAGACATCTTTTTTTT ........................................................................................................((((..(((.....)))..)))).....(((.....(((((((((....((((((.....))))))............)))))))))....))).................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|