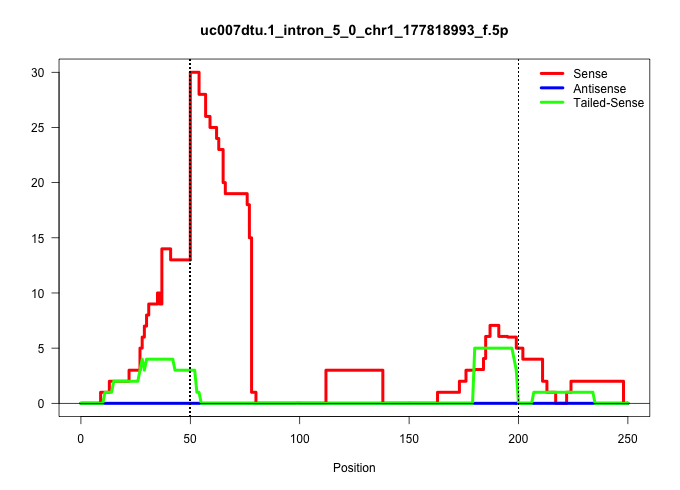
| Gene: Exo1 | ID: uc007dtu.1_intron_5_0_chr1_177818993_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(8) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| AGGCTGTCATCACAGAGGACTCTGACCTCCTCGCATTTGGCTGTAAGAAGGTATTGTACTGCACCTGTTACACATGGCGCGGTATGGGGGAGGATTGTGGGTGCTGAGGAGCTAAAATCTTCGTTTTATGCTTCTGTTTTTTTGTTTTAAATAGTGTTCATTTTTGGTTTTGCTAATTGATGCTTTAGGCTCTGTGTTCCTTCAGGTTGAAGGGCTTGGTTTTATCCTTTGGAGTTTGGTTTGGTTTGAT ..................................................((((.(((((((.((((.....)).)).)))))))....(((((((...(((........)))..))))))).....))))....................................................................................................................... ..................................................51.........................................................................................142.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTATTGTACTGCACCTGTTACACATGGC............................................................................................................................................................................ | 28 | 1 | 15.00 | 15.00 | 6.00 | 5.00 | - | - | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGCTTTAGGCTCTGTGacaa.................................................. | 20 | acaa | 3.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGGCTGTAAGAAGGTATTGTACTGCACC......................................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATTGTACTGCACCTGTTACACATGG............................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TAAAATCTTCGTTTTATGCTTCTGTT................................................................................................................ | 26 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................TAGGCTCTGTGTTCCTTCAGGTTGAA....................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGCTTTAGGCTCTGTGaaa................................................... | 19 | aaa | 2.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........ATCACAGAGGACTCTGACC............................................................................................................................................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TCGCATTTGGCTGTAAGAAGGTATTGT................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TATCCTTTGGAGTTTGGTTTcgtt.... | 24 | cgtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................CCTCGCATTTGGCTGTAAGAAGGTt..................................................................................................................................................................................................... | 25 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGCTTTAGGCTCTGTGaaaa.................................................. | 20 | aaaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............AGAGGACTCTGACCTCCTCGCATTTGGC................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TTTGGCTGTAAGAAGGTATTGTACTGC............................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTATTGTACTGCACCTGTTACACATG.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................TTGATGCTTTAGGCTCTGTGTTCCTT................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............AGGACTCTGACCTCCTCGCATTTGGCTa............................................................................................................................................................................................................... | 28 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCCTTTGGAGTTTGGTTTGGTTTG.. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................TCGCATTTGGCTGTAAGAAGGTgat................................................................................................................................................................................................... | 25 | gat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................TTGTACTGCACCTGTTACACATGGCGC.......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................TGAAGGGCTTGGTTTTATCCTTTGGAGa............... | 28 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................GGCTCTGTGTTCCTTCAGGTTGAAGGGCTT................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................TGGCTGTAAGAAGGTATTGTACTGCACCT........................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TTAGGCTCTGTGTTCCTTCAGGTTGAAGG..................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTGGTTTTGCTAATTGATGCTTTAGGCT........................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................TAATTGATGCTTTAGGCTCTGTGTTC................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCTCGCATTTGGCTGTAAGAAGGTA..................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGCTTTAGGCTCTGTGaa.................................................... | 18 | aa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TCACAGAGGACTCTGACCTCCTCGCAT...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................CGCATTTGGCTGTAAGAAGGTATTGTAC............................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................TGGCTGTAAGAAGGTATTGTACTGCA........................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCTCGCATTTGGCTGTAAGAAGGga..................................................................................................................................................................................................... | 26 | ga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TATCCTTTGGAGTTTGGTTTGGTTTG.. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGACCTCCTCGCATTTGGCTGTAAGAAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................CCTCGCATTTGGCTGTAAGAAGGTAT.................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCTCGCATTTGGCTGTAAGAAGGTAT.................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........ACAGAGGACTCTGAggca............................................................................................................................................................................................................................. | 18 | ggca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................CTCGCATTTGGCTGTAAGAAGGTATTGT................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................................TGCTTTAGGCTCTGT....................................................... | 15 | 16 | 0.06 | 0.06 | - | - | - | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGCTGTCATCACAGAGGACTCTGACCTCCTCGCATTTGGCTGTAAGAAGGTATTGTACTGCACCTGTTACACATGGCGCGGTATGGGGGAGGATTGTGGGTGCTGAGGAGCTAAAATCTTCGTTTTATGCTTCTGTTTTTTTGTTTTAAATAGTGTTCATTTTTGGTTTTGCTAATTGATGCTTTAGGCTCTGTGTTCCTTCAGGTTGAAGGGCTTGGTTTTATCCTTTGGAGTTTGGTTTGGTTTGAT ..................................................((((.(((((((.((((.....)).)).)))))))....(((((((...(((........)))..))))))).....))))....................................................................................................................... ..................................................51.........................................................................................142.......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|