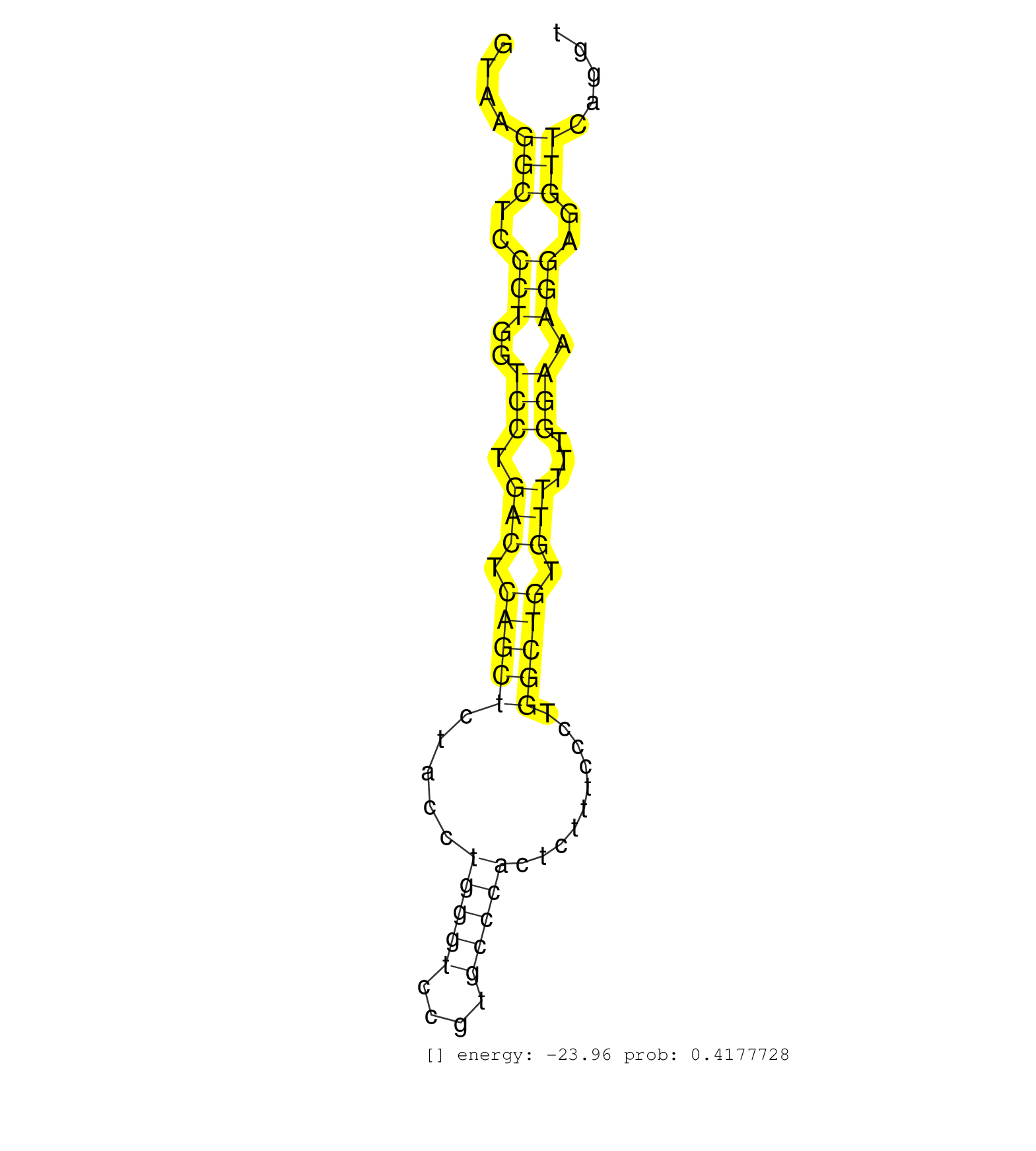

| Gene: Igsf9 | ID: uc007dqn.1_intron_10_0_chr1_174424346_f | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| TCCCGTGCTCCGCCCGGGGAGACCCTCCTCCTATTGTCTCTTGGGCCAAGGTAAGGCTCCCTGGTCCTGACTCAGCTCTACCTGGGTCCGTGCCCACTCTTTCCCTGGCTGTGTTTTTGGAAAGGAGGTTCAGGTTTTCACAGGCAACATGGGGAGGGGCACAGGTTACTTGGAGAGATGAGTGGCCCCCTTGGTGAGGGGTGGCTGCCTGTGTCTTCAGGGCTGACCAGTGGTTCTGTAGGTGGGCCGGGGGCTGCAGGGCCAGGCCCAGGTGGACAGCAACAACAGCCT ......................................................(((..(((..(((.(((.(((((.....(((((....)))))..........))))).)))...))).)))..)))................................................................................................................................................................. ..................................................51..................................................................................135.......................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TGGCTGTGTTTTTGGAAAGGAGGTTC................................................................................................................................................................ | 26 | 1 | 12.00 | 12.00 | 1.00 | - | 1.00 | 1.00 | 3.00 | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAGGCTCCCTGGTCCTGACTCAGC....................................................................................................................................................................................................................... | 26 | 1 | 12.00 | 12.00 | 8.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| .........................................................................................................TGGCTGTGTTTTTGGAAAGGAGGTT................................................................................................................................................................. | 25 | 1 | 6.00 | 6.00 | - | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................TTCCCTGGCTGTGTTTTTGGAAAGGAGG................................................................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 1.00 | - | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..........................................................................................................................................................................................................................AGGGCTGACCAGTGGTTCTGTAGa................................................. | 24 | a | 4.00 | 0.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................CACAGGTTACTTGGAGAGATGAGTG........................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................TGGCTGTGTTTTTGGAAAGGAGGT.................................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGCTCCCTGGTCCTGACTCAGCTa..................................................................................................................................................................................................................... | 28 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TTCAGGTTTTCACAGGCAACATGGGG......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................GGTGGACAGCAACAAagt... | 18 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGGCTGTGTTTTTGGAAAGGAGGTa................................................................................................................................................................. | 25 | a | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCCTGGCTGTGTTTTTGGAAAGGAGGTT................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................TGGCTGTGTTTTTGGAAAGGAGGTTta............................................................................................................................................................... | 27 | ta | 1.00 | 6.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGGCTCCCTGGTCCTGACTCAGCT...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCCTGGCTGTGTTTTTGGAAAGGAGG................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TGGCTGTGTTTTTGGAAAGGAGGTTCAGG............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TCCCTGGCTGTGTTTTTGGAAAGGAGG................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTTCCCTGGCTGTGTTTTTGGAAAGGA..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TGGCTGTGTTTTTGGAAAGGAG.................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCCCGTGCTCCGCCCGGGGAGACCCTCCTCCTATTGTCTCTTGGGCCAAGGTAAGGCTCCCTGGTCCTGACTCAGCTCTACCTGGGTCCGTGCCCACTCTTTCCCTGGCTGTGTTTTTGGAAAGGAGGTTCAGGTTTTCACAGGCAACATGGGGAGGGGCACAGGTTACTTGGAGAGATGAGTGGCCCCCTTGGTGAGGGGTGGCTGCCTGTGTCTTCAGGGCTGACCAGTGGTTCTGTAGGTGGGCCGGGGGCTGCAGGGCCAGGCCCAGGTGGACAGCAACAACAGCCT ......................................................(((..(((..(((.(((.(((((.....(((((....)))))..........))))).)))...))).)))..)))................................................................................................................................................................. ..................................................51..................................................................................135.......................................................................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................TGTCTTCAGGGCTGACCAGTGGTTCTGT.................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................CTCCCTGGTCCTGACTCAGCTCTAa................................................................................................................................................................................................................... | 25 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................CCACTCTTTCCCTGcttt........................................................................................................................................................................................ | 18 | cttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................................................TCTTCAGGGCTGACCAGTGGTTCTGTa.................................................... | 27 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TTTTGGAAAGGAGGTTCAGGTTTTCA....................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................GCAACAACAGCCTgca | 16 | gca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |