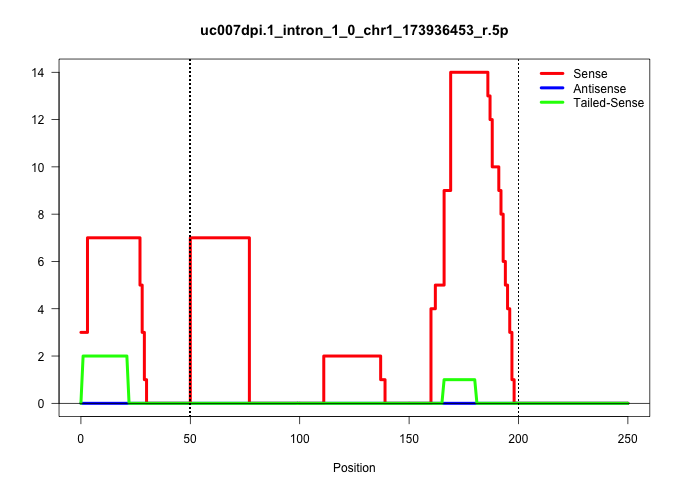
| Gene: Vangl2 | ID: uc007dpi.1_intron_1_0_chr1_173936453_r.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| TACTACGAGGAAGCCGAGCATGAGCGCAGAGTGCGCAAGCGCAGGGCCAGGTGGGTGCTGGCTGTGTGAGTCCGGGGCGGGGCGGGCACCTGAGGGAGGGGAAAAGCCAACTGAGTGGTACCCGTGGGATGAACTGGGCTGAGAAAGCTCAGGAGACCCCTGTACCTGATGAGAACGGAACAGTGATGATACAGGGTTTGTATATTCATTTCCCAGTGCTTCCGTGACTGATCACCACGGACTCTCAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGGTGCTGGCTGTGTGAGTCCGGGG............................................................................................................................................................................. | 27 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .ACTACGAGGAAGCCGAGCATt.................................................................................................................................................................................................................................... | 21 | t | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TGATGAGAACGGAACAGTGATGATACA......................................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGAGAACGGAACAGTGATGATACAGGGT..................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...TACGAGGAAGCCGAGCATGAGCGCAG............................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................TGAGAACGGAACAGTGATGATACAGG....................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................TGTACCTGATGAGAACGGAACAGTGATGATA........................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................................................................................................TGTACCTGATGAGAACGGAACAGTGA................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................TACCTGATGAGAACGGAACAGTGATG.............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGAGAACGGAACAGTGATGATACAGGGTT.................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................TGAGTGGTACCCGTGGGATGAACTGG................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................TGATGAGAACGGtgt..................................................................... | 15 | tgt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGTACCTGATGAGAACGGAACAGTGATG.............................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................................TGAGTGGTACCCGTGGGATGAACTGGGC............................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TGATGAGAACGGAACAGTGATGATAC.......................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...TACGAGGAAGCCGAGCATGAGCGCA.............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...TACGAGGAAGCCGAGCATGAGCGCAGA............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGTACCTGATGAGAACGGAACAGTGAT............................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TGATGAGAACGGAACAGTGATGATACAG........................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................TGAGAACGGAACAGTGATGATACAGGG...................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| TACTACGAGGAAGCCGAGCATGAGCGCAGAGTGCGCAAGCGCAGGGCCAGGTGGGTGCTGGCTGTGTGAGTCCGGGGCGGGGCGGGCACCTGAGGGAGGGGAAAAGCCAACTGAGTGGTACCCGTGGGATGAACTGGGCTGAGAAAGCTCAGGAGACCCCTGTACCTGATGAGAACGGAACAGTGATGATACAGGGTTTGTATATTCATTTCCCAGTGCTTCCGTGACTGATCACCACGGACTCTCAGCA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|