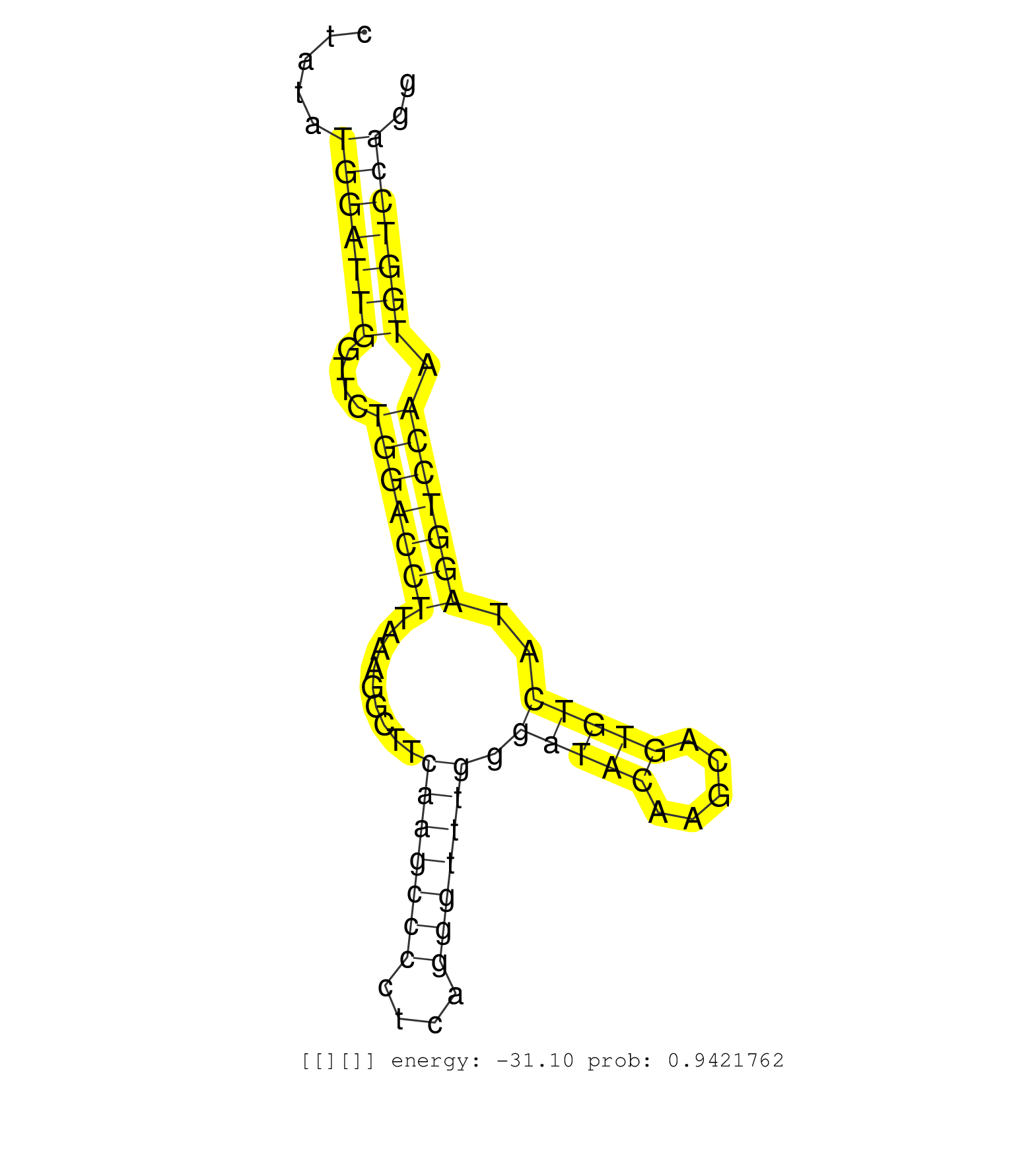

| Gene: Sacy | ID: uc007dje.1_intron_19_0_chr1_167482309_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| TTTGGATATGGACACTGTCAAGAGGATGGTGACATCCCAAGGATTTAAAAGTAAAACTTCACTTCTAACAAGTCCCAACTCCAGCTGGGAAATCTTAGGAGACTGAGGTCTATATGGATTGGTTCTGGACCTTAAAGGCTTCAAGCCCCTCAGGGTTTGGGATACAAGCAGTGTCATAGGTCCAATGGTCCAGGGAAGAGCCAGCAATGCACCTGGGGTATCAGAGTCTAGCTCACTGTGATTTCCACAC ..................................................................................................................(((((((....(((((((.........(((((((....))))))).(((((.....)))))..))))))).))))))).......................................................... .............................................................................................................110.................................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................TGGATTGGTTCTGGACCTTAAAGGCTT............................................................................................................. | 27 | 1 | 8.00 | 8.00 | 5.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..TGGATATGGACACTGTCAAGAGGATG.............................................................................................................................................................................................................................. | 26 | 1 | 8.00 | 8.00 | 3.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TATGGATTGGTTCTGGACCTTAAAGGC............................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................TGGATTGGTTCTGGACCTTAAAGGCT.............................................................................................................. | 26 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TAGGAGACTGAGGTCTATATGGATTG................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .TTGGATATGGACACTGTCAAGAGGATG.............................................................................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TATGGATTGGTTCTGGACCTTAAAGGCT.............................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..TGGATATGGACACTGTCAAGAGGATGG............................................................................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGGATTGGTTCTGGACCTTAAAGGCTTC............................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AATGCACCTGGGGTtcag........................... | 18 | tcag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................................CCAGCTGGGAAATCTTAGGAGACTGAGG.............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........TGGACACTGTCAAGAGGATGGTGACATC...................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................TGGGAAATCTTAGGAGACTGAGGTCT........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........TGGACACTGTCAAGAGGATGGTGACATCCC.................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........TGGACACTGTCAAGAGGATGGTGACAT....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TATGGATTGGTTCTGGACCTTAAAGG................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TACAAGCAGTGTCATAGGTCCAATGGTt............................................................ | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................TGGATTGGTTCTGGACCTTAAAGGC............................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................TCAGGGTTTGGGATACAAGCAGTGTC........................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TGCACCTGGGGTATCAGAGTCTAGCTCA............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................TAGGAGACTGAGGaacg.......................................................................................................................................... | 17 | aacg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................CTATATGGATTGGTTCTGGACCTTAAA.................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .TTGGATATGGACACTGTCAAGAGGAT............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................TCAAGAGGATGGTGgtt........................................................................................................................................................................................................................ | 17 | gtt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................CTATATGGATTGGTTCTGGACCTTAA................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TATATGGATTGGTTCTGGACCTTAAAGGC............................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TATGGATTGGTTCTGGACCTTAAAGGCTT............................................................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TGGATATGGACACTGTCAAGAGGAcggt............................................................................................................................................................................................................................ | 28 | cggt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GGATTGGTTCTGGACCTTAAAGGCTTC............................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| TTTGGATATGGACACTGTCAAGAGGATGGTGACATCCCAAGGATTTAAAAGTAAAACTTCACTTCTAACAAGTCCCAACTCCAGCTGGGAAATCTTAGGAGACTGAGGTCTATATGGATTGGTTCTGGACCTTAAAGGCTTCAAGCCCCTCAGGGTTTGGGATACAAGCAGTGTCATAGGTCCAATGGTCCAGGGAAGAGCCAGCAATGCACCTGGGGTATCAGAGTCTAGCTCACTGTGATTTCCACAC ..................................................................................................................(((((((....(((((((.........(((((((....))))))).(((((.....)))))..))))))).))))))).......................................................... .............................................................................................................110.................................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|