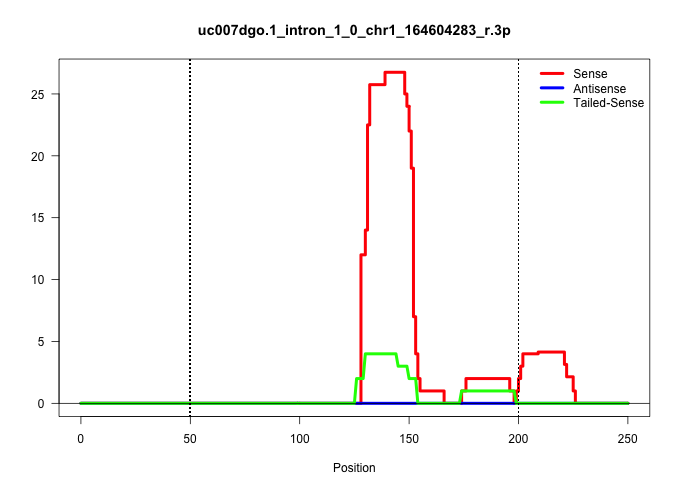
| Gene: Bat2d | ID: uc007dgo.1_intron_1_0_chr1_164604283_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(2) PIWI.mut |
(21) TESTES |
| ATTATATGCAAAAGGAGTAGAAAGTTAAGTTTAATTGGATATAGTATAGCACTATAATTACAGGTTAATGTAAGTATATTCTTTGGGGCACATTGTCATTGAAACGGATTATTTACCAGCTCAGAATGTGGTAGGAGCTATCAGAACTTAGTGATCAAGTGAAGTCGTAGTTACTAATTTCTGATGCTCTTCCCCTGCAGAAGAGAGCTGTGGGAAGAGGACTCAGCCACAAGACATTTGGTCCTAGGTG ..............................................................................................................................(((((.((((((.(((((((.((((((((....((....))..)))))))).))))))))))))).))..)))................................................... ...........................................................................................................................124.........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................TGGTAGGAGCTATCAGAACTTAGT.................................................................................................. | 24 | 1 | 11.00 | 11.00 | 2.00 | - | - | 3.00 | 1.00 | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................TAGGAGCTATCAGAACTTAGT.................................................................................................. | 21 | 1 | 10.00 | 10.00 | 3.00 | 1.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGGTAGGAGCTATCAGAACTTAG................................................................................................... | 23 | 1 | 8.00 | 8.00 | - | 1.00 | 1.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ...................................................................................................................................TAGGAGCTATCAGAACTTAGTG................................................................................................. | 22 | 1 | 4.00 | 4.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TAGGAGCTATCAGAACTTAG................................................................................................... | 20 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GTAGGAGCTATCAGAACTTAGTGt................................................................................................ | 24 | t | 2.00 | 0.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGGTAGGAGCTATCAGAACTTA.................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................TAATTTCTGATGCTCTTCCCCT...................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................TAGGAGCTATCAGAACTTAGTGA................................................................................................ | 23 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................AGGAGCTATCAGAACTTAGTG................................................................................................. | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GTAGGAGCTATCAGAACTTAGT.................................................................................................. | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CAAGTGAAGTCGTAGTTACT........................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GAAGAGAGCTGTGGGAAGAGGACTCAG........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GTAGGAGCTATCAGAACTTA.................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GAGAGCTGTGGGAAGAGGACTCA......................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGGTAGGAGCTATCAGAACTT..................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................TGTGGTAGGAGCTATCAGAACagt.................................................................................................... | 24 | agt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................AGGAGCTATCAGAACTTAGTGA................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ATCAGAACTTAGTGATCAAGTGAAGTC.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........AAGGAGTAGAAAGTTAAGT............................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGTGGTAGGAGCTATCcga......................................................................................................... | 19 | cga | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGGTAGGAGCTATCAGAACTTAGTG................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................................................................................................................AGAGAGCTGTGGGAAGAGGA............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GAGAGCTGTGGGAAGAGGACTC.......................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AAGAGAGCTGTGGGAAGAGGAC............................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TGGTAGGAGCTATCAGAACT...................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TAATTTCTGATGCTCTTCCCCTGaa................................................... | 25 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGTGGTAGGAGCTATCAGAACTTAG................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TAGGAGCTATCAGAACTTAGTGAT............................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ATTTCTGATGCTCTTCCCCTGC.................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TAGGAGCTATCAGAACT...................................................................................................... | 17 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................AGGAGCTATCAGAACT...................................................................................................... | 16 | 4 | 0.25 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GTGGGAAGAGGACTCA......................... | 16 | 7 | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATTATATGCAAAAGGAGTAGAAAGTTAAGTTTAATTGGATATAGTATAGCACTATAATTACAGGTTAATGTAAGTATATTCTTTGGGGCACATTGTCATTGAAACGGATTATTTACCAGCTCAGAATGTGGTAGGAGCTATCAGAACTTAGTGATCAAGTGAAGTCGTAGTTACTAATTTCTGATGCTCTTCCCCTGCAGAAGAGAGCTGTGGGAAGAGGACTCAGCCACAAGACATTTGGTCCTAGGTG ..............................................................................................................................(((((.((((((.(((((((.((((((((....((....))..)))))))).))))))))))))).))..)))................................................... ...........................................................................................................................124.........................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|