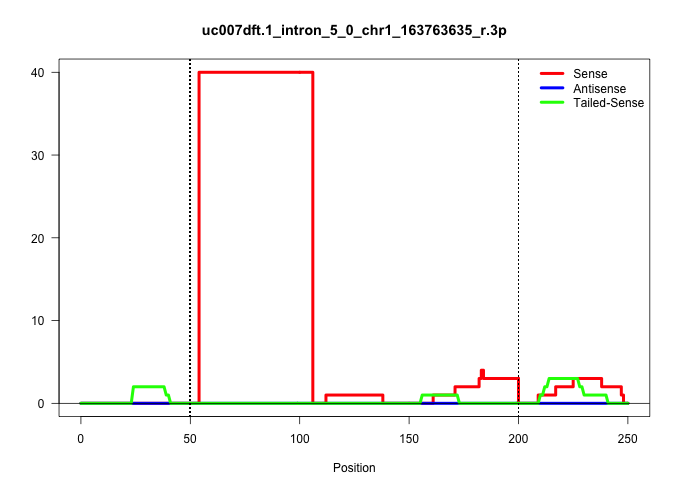
| Gene: AI848100 | ID: uc007dft.1_intron_5_0_chr1_163763635_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(13) TESTES |
| TAGCGTGCCCAACTTCATTTCCAGGATCCTTGATGTCCAGGATCTCTCTATTTGGTTATACCTTTTATGTGTGTTACTGGATTGCACAGTCCTGAATTGGTCATTTAAGAAAAAGCAGGATTGTTCAGTCTCAAAATCAGAATGATTTTCCAGTACATAATGAACGAGAAAATGGATTGAAAGCAACTCTTACTTTGTAGGTACCGCAAACAAATGGAAGAAATGCAGAAGGCTTTCAATAAGACAATAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................GTTATACCTTTTATGTGTGTTACTGGATTGCACAGTCCTGAATTGGTCATTT................................................................................................................................................ | 52 | 1 | 40.00 | 40.00 | 40.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................ATGGATTGAAAGCAACTCTTACTTTGTAG.................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................GATCCTTGATGTCgg................................................................................................................................................................................................................... | 15 | gg | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................CAGAAGGCTTTCAATAAGACAA... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................CAACTCTTACTTTGTAG.................................................. | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................GAACGAGAAAATGGATTGAAAGC.................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................ATAATGAACGAGActgt............................................................................. | 17 | ctgt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................GATCCTTGATGTCggct................................................................................................................................................................................................................. | 17 | ggct | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................AATGGAAGAAATGCgtgt.................... | 18 | gtgt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................GCAACTCTTACTTTGTAG.................................................. | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................ACAAATGGAAGAAATGCAGAAGGCTTTCA............ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................AAGCAGGATTGTTCAGTCTCAAAATC................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................AAGAAATGCAGAAGGCTTTCAATAAGACAAT.. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................TGGAAGAAATGCAGAAGGCTTTCAAat......... | 27 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................CAAATGGAAGAAATGgct...................... | 18 | gct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| TAGCGTGCCCAACTTCATTTCCAGGATCCTTGATGTCCAGGATCTCTCTATTTGGTTATACCTTTTATGTGTGTTACTGGATTGCACAGTCCTGAATTGGTCATTTAAGAAAAAGCAGGATTGTTCAGTCTCAAAATCAGAATGATTTTCCAGTACATAATGAACGAGAAAATGGATTGAAAGCAACTCTTACTTTGTAGGTACCGCAAACAAATGGAAGAAATGCAGAAGGCTTTCAATAAGACAATAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................CCAGGATCTCTCTATcag....................................................................................................................................................................................................... | 18 | cag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |