
| Gene: Rnasel | ID: uc007dag.1_intron_0_0_chr1_155596738_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(15) TESTES |
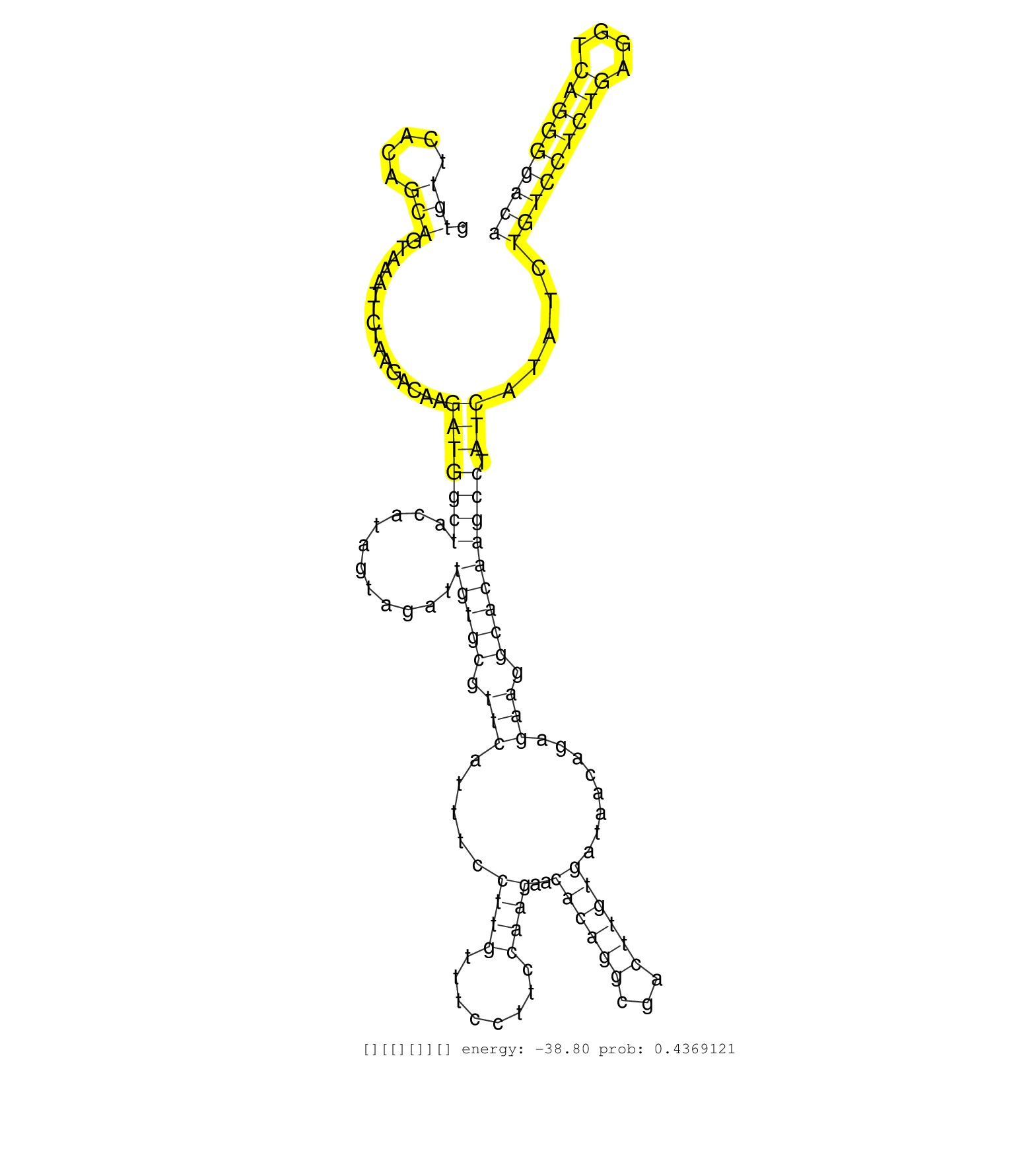
| TCTGAACCTGTAAGCCAGTCCTAATTAAATGTCCTTATAAGACTTGCCTTGGTCATGGTGTGTGTTCACAGCAGTAAATTCTAAGACAAGATGGCTACATAGTAGATTGTGCGTTCATTTCCTTGTTTCCTTCCAAGAACACAGGCGACTTGTGATAACAGAGAAGGCACAAGCCTATCATATCTGTCCTCTGAGGTCAGGGGACAGCCCTTGTCTTCCTCCAGGAGGTGAAGATTGCTCAGGCTGTGCC ..............................................................(((.....)))................(((((((...........(((((.(((.....((((........))))..((((((...))))))........))).))))))))).))).....(((((((((....)))))))))............................................ .............................................................62..............................................................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................CACAGCAGTAAATTCTAAGACAAGATG............................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TAGTAGATTGTGCGTTCATTTCCTTG............................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................ACAGCAGTAAATTCTAAGACAAGATGGC........................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................CACAGCAGTAAATTCTAAGACAAGATGGCT.......................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TATCATATCTGTCCTCTGAGGTCAGGG................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................AGAGAAGGCACAAGCCTggga...................................................................... | 21 | ggga | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GTTCACAGCAGTAAATTCTAAGACAAGATGGC........................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................CAGTAAATTCTAAGACAAGATGGCTACAT...................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................CAGCAGTAAATTCTAAGACAAGATGGCTAC........................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TCCAGGAGGTGAAGATTGCTCAGGCTGTGC. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TAAGACAAGATGGCTACATAGTAGATT.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGTGTTCACAGCAGTAAATTCTAAGACA.................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TCTAAGACAAGATGGCTACATAGTAGA................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................TATAAGACTTGCCTTGGTCATGGat.............................................................................................................................................................................................. | 25 | at | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................CAGCAGTAAATTCTAAGACAAGATGGCT.......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................TTCACAGCAGTAAATTCTAAGACAAGATG............................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................GTAAATTCTAAGACAAGATGGCT.......................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGTGTTCACAGCAGTAAATTCTAAGAC................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................................TACATAGTAGATTGTGCGTTCATTTC................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TAGATTGTGCGTTCATTTCCTTGTT........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................ACAGCAGTAAATTCTAAGACAAGATGGCT.......................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ACACAGGCGACTTGTGATAACAGAGA...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TCTTCCTCCAGGAGGTGAAGATTGCTC.......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................TAAATGTCCTTATAAGACTTGCCTTGGTCA................................................................................................................................................................................................... | 30 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| TCTGAACCTGTAAGCCAGTCCTAATTAAATGTCCTTATAAGACTTGCCTTGGTCATGGTGTGTGTTCACAGCAGTAAATTCTAAGACAAGATGGCTACATAGTAGATTGTGCGTTCATTTCCTTGTTTCCTTCCAAGAACACAGGCGACTTGTGATAACAGAGAAGGCACAAGCCTATCATATCTGTCCTCTGAGGTCAGGGGACAGCCCTTGTCTTCCTCCAGGAGGTGAAGATTGCTCAGGCTGTGCC ..............................................................(((.....)))................(((((((...........(((((.(((.....((((........))))..((((((...))))))........))).))))))))).))).....(((((((((....)))))))))............................................ .............................................................62..............................................................................................................................................206.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................................GGGGACAGCCCTTGTCTTCCTC............................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................ATATCTGTCCTCTGAttct........................................................ | 19 | ttct | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GGGACAGCCCTTGTCTTCCTCCAGGAGGTG.................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........TAAGCCAGTCCTAATTAAATGTCCTTAT.................................................................................................................................................................................................................... | 28 | 3 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |