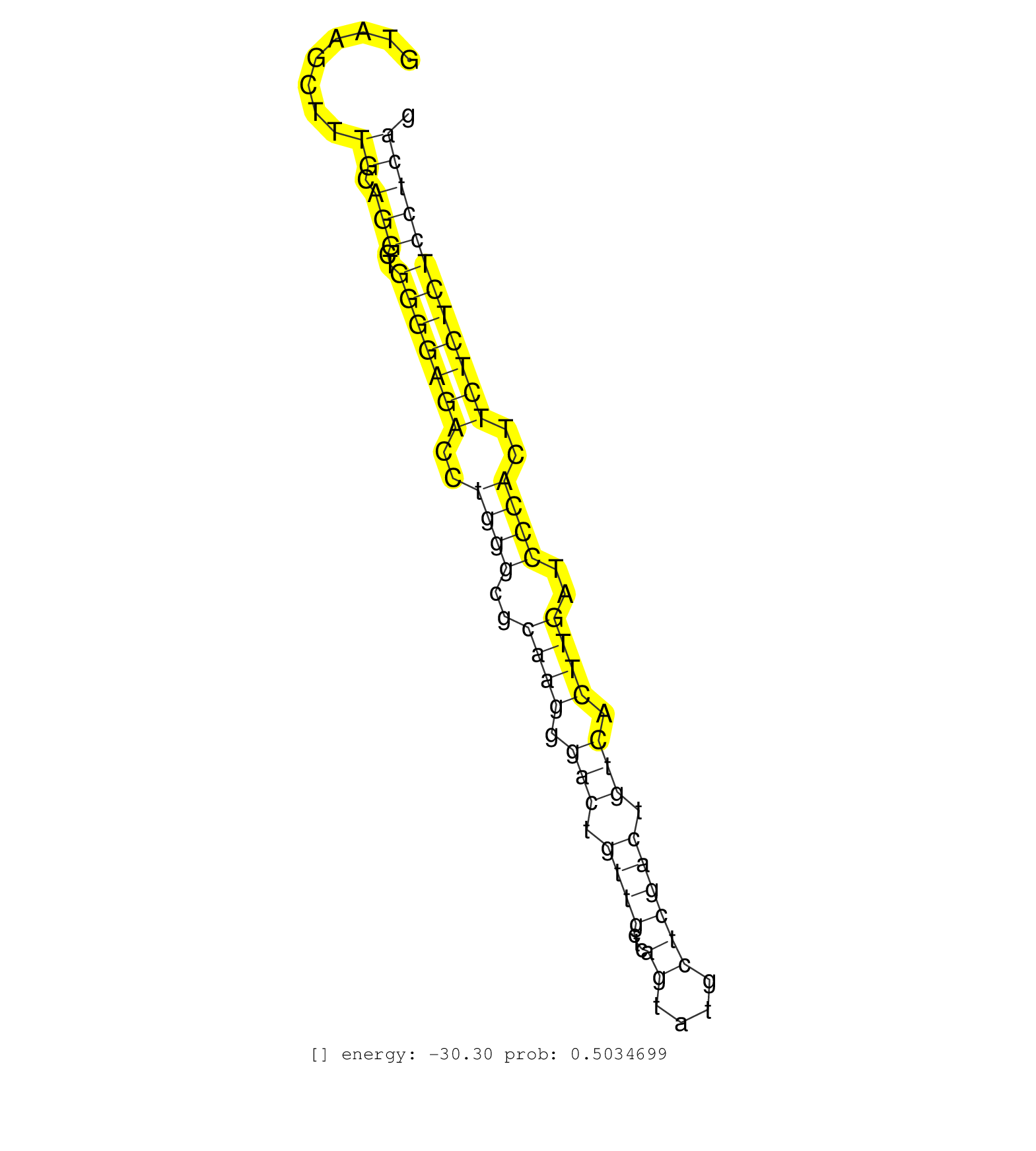

| Gene: Mapkapk2 | ID: uc007cmv.1_intron_1_0_chr1_132952417_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
| CCACCCAGAGAATGACCATCACAGAATTCATGAACCACCCCTGGATCATGGTAAGCTTTGCAGGCTGGGGAGACCTGGGCGCAAGGGACTGTTGCTCAGTATGCTCGACTGTCACTTGATCCCACTTCTCTCTCCTCAGCAATCTACGAAGGTCCCTCAGACTCCACTGCACACCAGCCGTGTCCTGAA ..........................................................((.(((..(((((((..((((..((((.(((.((((...((....)))))).))).))))..))))..))))))))))))................................................... ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGCTTTGCAGGCTGGGGAGACC.................................................................................................................. | 25 | 1 | 14.00 | 14.00 | 8.00 | 4.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| ..................................................GTAAGCTTTGCAGGCTGGGGAGAC................................................................................................................... | 24 | 1 | 12.00 | 12.00 | 6.00 | 5.00 | - | 1.00 | - | - | - | - | - | - |
| .......GAGAATGACCATCACAGAATTCATGAACCA........................................................................................................................................................ | 30 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCTTTGCAGGCTGGGGAGACt.................................................................................................................. | 25 | t | 1.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........ATGACCATCACAGAATTCATGAACCACCCCTG.................................................................................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................CACTTGATCCCACTTCTCTCT........................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTAAGCTTTGCAGGCTGGGGAGA.................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ............TGACCATCACAGAATTCATGAACCACCCC.................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................GCAGGCTGGGGAGACCTGGGCGCAAGGGACTGT................................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ............TGACCATCACAGAATTCATGAACCACCC..................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .......GAGAATGACCATCACAGAATTCATGAACC......................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTAAGCTTTGCAGGCTGGGGAGACCT................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................CGAAGGTCCCTCAGACTCCACTGCACAC............... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTAAGCTTTGCAGGCTGGGGAGACCTatt.............................................................................................................. | 29 | att | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................TGAACCACCCCTGGA................................................................................................................................................ | 15 | 8 | 0.12 | 0.12 | - | - | - | 0.12 | - | - | - | - | - | - |
| CCACCCAGAGAATGACCATCACAGAATTCATGAACCACCCCTGGATCATGGTAAGCTTTGCAGGCTGGGGAGACCTGGGCGCAAGGGACTGTTGCTCAGTATGCTCGACTGTCACTTGATCCCACTTCTCTCTCCTCAGCAATCTACGAAGGTCCCTCAGACTCCACTGCACACCAGCCGTGTCCTGAA ..........................................................((.(((..(((((((..((((..((((.(((.((((...((....)))))).))).))))..))))..))))))))))))................................................... ..................................................51......................................................................................139................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................CTCGACTGTCACTgc......................................................................... | 15 | gc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - |