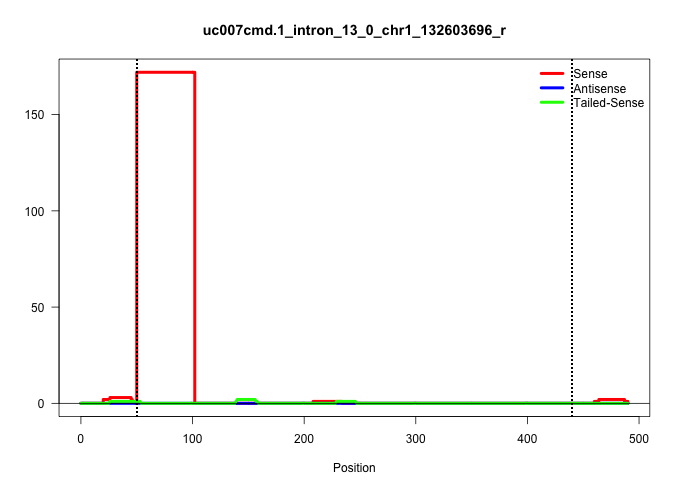
| Gene: Pfkfb2 | ID: uc007cmd.1_intron_13_0_chr1_132603696_r | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(9) TESTES |
| TCTTATGATTTCTTTCGACATGACAATGAAGAGGCTATGAAGATCCGCAAGTGAGTTTTTTTTTAAAGGCCTGACTGAAATCAACTGTTTTTAGGCAGTTGTGAGATTGAATGTGTTAGGGTCCTGTTTTTCTCAGGAAATATTTTGAGGGGATTTTAAGGGAAATTACTAGATTGAAAGACCAGCTTAGGTTGGATGGGTAGCAGACAGGACTCTAGAAGATGATTTTATGTGATCCTAAAGAGGTATAAGTCCTATGAATTGGACAGGGGGAAGTTGTAGGATGGTGTATGAACTCCATGGGAAAATTGGAAGATTTAGTTAAGGCAAACATTACAACCAGAAAGATACTGAGTATGAAGTGATTCCTACAACTGTGTTTATCTCTGGCCTTGTTTCATCTTAGCTCTTTTCACTGATTTTCTTTGTTCTCATGTCAGACAGTGTGCTCTGGTGGCACTGGAAGATGTTAAAGCCTATTTTACTGAAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGTTTTTTTTTAAAGGCCTGACTGAAATCAACTGTTTTTAGGCAGTTGT.................................................................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 172.00 | 172.00 | 172.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................AGATGTTAAAGCCTATTTTACTGAAG | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..........................TGAAGAGGCTATGAAGATCCGCAAacag.................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | acag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |
| ....................TGACAATGAAGAGGCTATGAAGATCCGCA......................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGAAGATGTTAAAGCCTATTTTACTG... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ....................TGACAATGAAGAGGCTATGAAGATC............................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................................................................................................................................................AGGACTCTAGAAGATGATTTTATGTG................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................ATATTTTGAGGGGATTgg............................................................................................................................................................................................................................................................................................................................................. | 18 | gg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................TATTTTGAGGGGATTgtat........................................................................................................................................................................................................................................................................................................................................... | 19 | gtat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................ATGTGATCCTAAAGtgaa................................................................................................................................................................................................................................................... | 18 | tgaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - |
| ..........................TGAAGAGGCTATGAAGATCC............................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| TCTTATGATTTCTTTCGACATGACAATGAAGAGGCTATGAAGATCCGCAAGTGAGTTTTTTTTTAAAGGCCTGACTGAAATCAACTGTTTTTAGGCAGTTGTGAGATTGAATGTGTTAGGGTCCTGTTTTTCTCAGGAAATATTTTGAGGGGATTTTAAGGGAAATTACTAGATTGAAAGACCAGCTTAGGTTGGATGGGTAGCAGACAGGACTCTAGAAGATGATTTTATGTGATCCTAAAGAGGTATAAGTCCTATGAATTGGACAGGGGGAAGTTGTAGGATGGTGTATGAACTCCATGGGAAAATTGGAAGATTTAGTTAAGGCAAACATTACAACCAGAAAGATACTGAGTATGAAGTGATTCCTACAACTGTGTTTATCTCTGGCCTTGTTTCATCTTAGCTCTTTTCACTGATTTTCTTTGTTCTCATGTCAGACAGTGTGCTCTGGTGGCACTGGAAGATGTTAAAGCCTATTTTACTGAAG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|