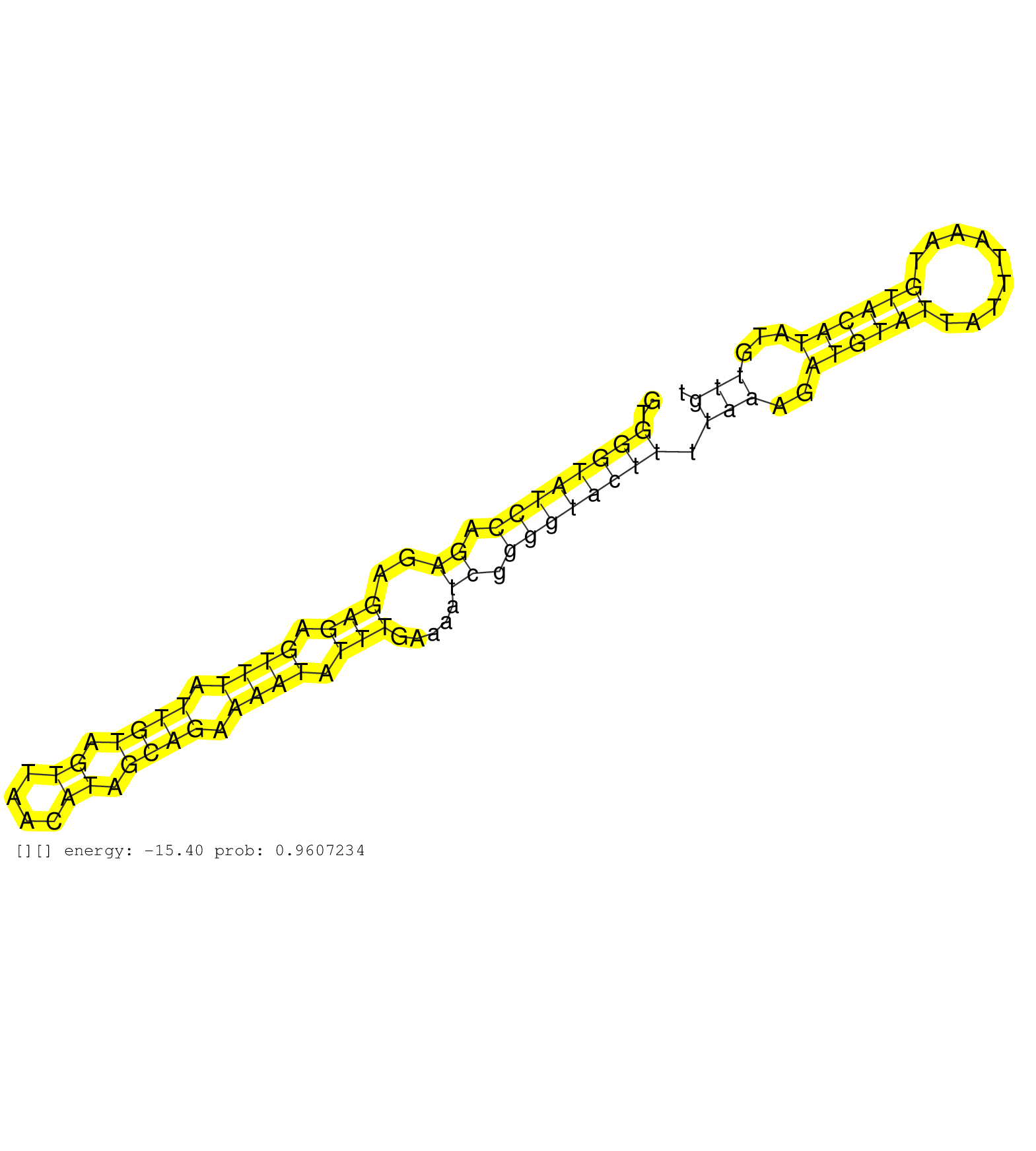
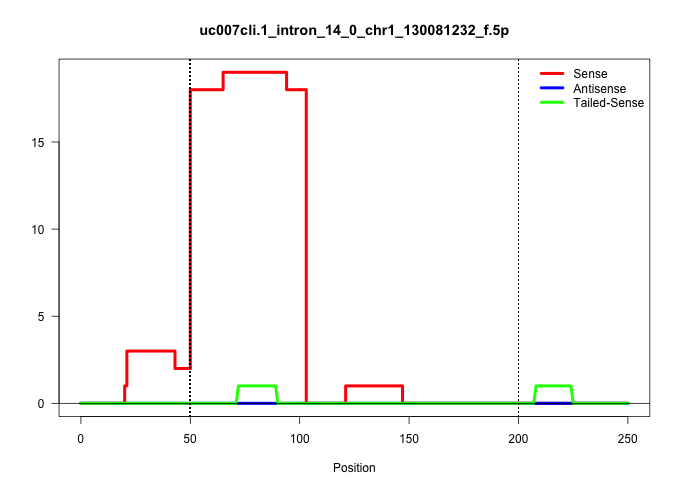
| Gene: R3hdm1 | ID: uc007cli.1_intron_14_0_chr1_130081232_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(9) TESTES |
| GGAAGCAACTGGCATACCACCTGGCAGTATTCTGATCAACCCACAAACAGGTGGGTATCCAGAGAGAGAGTTTATTGTAGTTAACATAGCAGAAAATATTTGAAAATCGGGGTACTTTTAAAGATGTATTATTTAAATGTACATATGTTGTTAGCTTTTTAATTTTGATAAATAATTGTCTTTGTCCTAAAAACTTTAATAGAAATGTTTAGGCTCCAAAATGTTAAAAACTATTTTTATCTTGTAAGAC ....................................................((((((((.((..(((.((((.((((.((....)).)))).)))).))).....)).)))))))).(((..((((((.........))))))...))).................................................................................................... ..................................................51..................................................................................................151................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGGTATCCAGAGAGAGAGTTTATTGTAGTTAACATAGCAGAAAATATTTG.................................................................................................................................................... | 52 | 1 | 18.00 | 18.00 | 18.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................AGATGTATTATTTAAATGTACATATG....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................TTAGGCTCCAAAAgtgc......................... | 17 | gtgc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................GAGAGTTTATTGTAGTTAACATAGCAGAA............................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .....................TGGCAGTATTCTGATCAACCCA............................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| .................CACCTGGCAGTATTCTGAT...................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ....................CTGGCAGTATTCTGATCAACCCACAAACAG........................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................TATTGTAGTTAACATgga................................................................................................................................................................ | 18 | gga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| .....................TGGCAGTATTCTGATCAACCCACAAACAG........................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| GGAAGCAACTGGCATACCACCTGGCAGTATTCTGATCAACCCACAAACAGGTGGGTATCCAGAGAGAGAGTTTATTGTAGTTAACATAGCAGAAAATATTTGAAAATCGGGGTACTTTTAAAGATGTATTATTTAAATGTACATATGTTGTTAGCTTTTTAATTTTGATAAATAATTGTCTTTGTCCTAAAAACTTTAATAGAAATGTTTAGGCTCCAAAATGTTAAAAACTATTTTTATCTTGTAAGAC ....................................................((((((((.((..(((.((((.((((.((....)).)))).)))).))).....)).)))))))).(((..((((((.........))))))...))).................................................................................................... ..................................................51..................................................................................................151................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................TCTTTGTCCTAAAAagta.......................................................... | 18 | agta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |