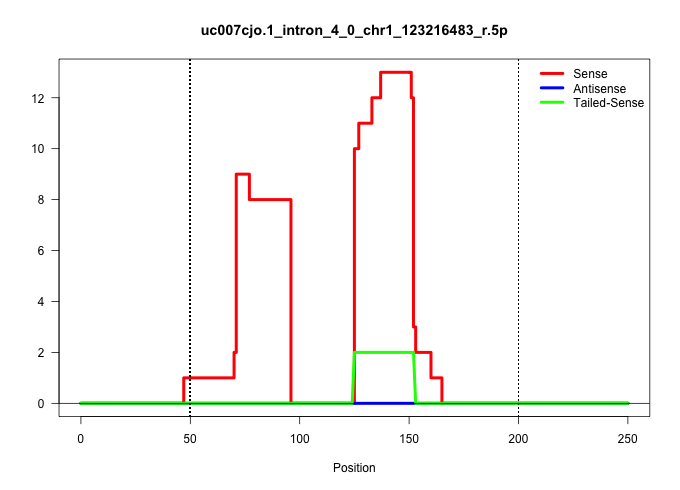
| Gene: Insig2 | ID: uc007cjo.1_intron_4_0_chr1_123216483_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(3) PIWI.mut |
(14) TESTES |
| CGCAGGTGGGGCCTGCTGGGGCGAGGGGTCCCCGGCTTACCTGGAGAGAGGTCAGCGAGATCTAGGGAAAAACCAGCGGGCTCATAGAGGGACAGGCTTTGGGGGATGCCGAGGTCCCTGGACAGTGGAAGACTGTCAGAGTGGAAGAAGGCTGGAGGACAGCGGAAAGGGCGACCCTGGAGGTCTGTTTGGGGACAGAGATGGGCCAGCTGTCTAGTGTACACCTGCCCGGACTCGGTGCCTTGTAGTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................TGGAAGACTGTCAGAGTGGAAGAAGGC.................................................................................................. | 27 | 1 | 9.00 | 9.00 | - | 3.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - | - |
| .......................................................................ACCAGCGGGCTCATAGAGGGACAGG.......................................................................................................................................................... | 25 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TGGAAGACTGTCAGAGTGGAAGAAGGCa................................................................................................. | 28 | a | 2.00 | 9.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGTCAGAGTGGAAGAAGGCTGGAGGAC.......................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................GAGGTCAGCGAGATCTAGGGAAAAACCAGC............................................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................AGAGTGGAAGAAGGCTGGAGGACAGCGG..................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................TGGAAGACTGTCAGAGTGGAAGAAGG................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................AACCAGCGGGCTCATAGAGGGACAGG.......................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................GAAGACTGTCAGAGTGGAAGAAGGCT................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| CGCAGGTGGGGCCTGCTGGGGCGAGGGGTCCCCGGCTTACCTGGAGAGAGGTCAGCGAGATCTAGGGAAAAACCAGCGGGCTCATAGAGGGACAGGCTTTGGGGGATGCCGAGGTCCCTGGACAGTGGAAGACTGTCAGAGTGGAAGAAGGCTGGAGGACAGCGGAAAGGGCGACCCTGGAGGTCTGTTTGGGGACAGAGATGGGCCAGCTGTCTAGTGTACACCTGCCCGGACTCGGTGCCTTGTAGTC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................TCAGCGAGATCTAaa.......................................................................................................................................................................................... | 15 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |