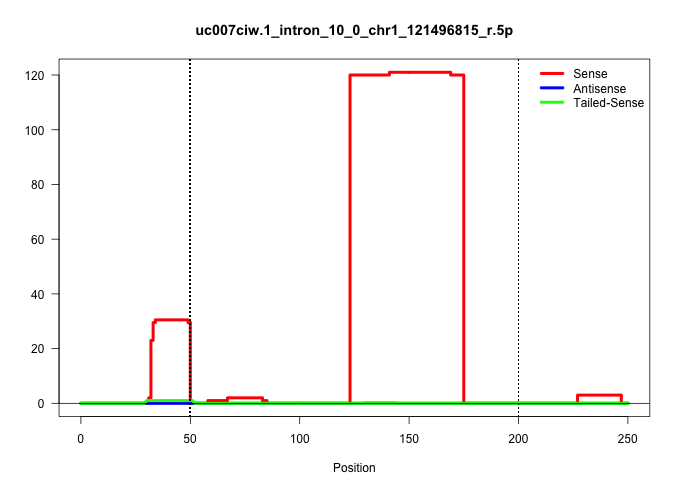
| Gene: Epb4.1l5 | ID: uc007ciw.1_intron_10_0_chr1_121496815_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CTCTTCTCTTCTCTTCTGTCAGCCAGCACAACACAACCTGAAGATCTTGGGTAAGTACTGCCTTGTAATGTAGGAACTGAGAAGATTTATATAGTTGCAATTGTTAATTAAACAAATATTTGTTGAAGGCCTTGCAAAATGTCAAATTTTCACAGGCATTAGGATTATAAGAAACAAAAACACACCAGATGAATTAATATTCAGAAAAATGAAGCTGTTTACAGAAGTAGAATAGGGTAGGAACAGCTAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................TGAAGGCCTTGCAAAATGTCAAATTTTCACAGGCATTAGGATTATAAGAAAC........................................................................... | 52 | 1 | 120.00 | 120.00 | 120.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................ACAACCTGAAGATCTTGG........................................................................................................................................................................................................ | 18 | 1 | 20.00 | 20.00 | - | 8.00 | 4.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - |
| .................................CAACCTGAAGATCTTGG........................................................................................................................................................................................................ | 17 | 2 | 6.50 | 6.50 | - | 3.00 | - | - | 0.50 | - | 1.00 | - | 1.50 | - | - | - | - | 0.50 |
| ...................................................................................................................................................................................................................................TAGAATAGGGTAGGAACAGC... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - |
| ...............................CACAACCTGAAGATCTTGG........................................................................................................................................................................................................ | 19 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TCAAATTTTCACAGGCATTAGGATTATA................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................ACAACCTGAAGATCTTG......................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGCCTTGTAATGTAGGAACTGAGAA....................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................ACACAACCTGAAGATCTTGGtg...................................................................................................................................................................................................... | 22 | tg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................AACCTGAAGATCTTGG........................................................................................................................................................................................................ | 16 | 4 | 1.00 | 1.00 | - | 0.25 | - | - | - | - | 0.50 | - | - | 0.25 | - | - | - | - |
| ...................................................................ATGTAGGAACTGAGAAGA..................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CTCTTCTCTTCTCTTCTGTCAGCCAGCACAACACAACCTGAAGATCTTGGGTAAGTACTGCCTTGTAATGTAGGAACTGAGAAGATTTATATAGTTGCAATTGTTAATTAAACAAATATTTGTTGAAGGCCTTGCAAAATGTCAAATTTTCACAGGCATTAGGATTATAAGAAACAAAAACACACCAGATGAATTAATATTCAGAAAAATGAAGCTGTTTACAGAAGTAGAATAGGGTAGGAACAGCTAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............GTCAGCCAGCACAAgggc........................................................................................................................................................................................................................... | 18 | gggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................CTTGCAAAATGTCAA......................................................................................................... | 15 | 15 | 0.07 | 0.07 | - | - | - | - | - | - | 0.07 | - | - | - | - | - | - | - |