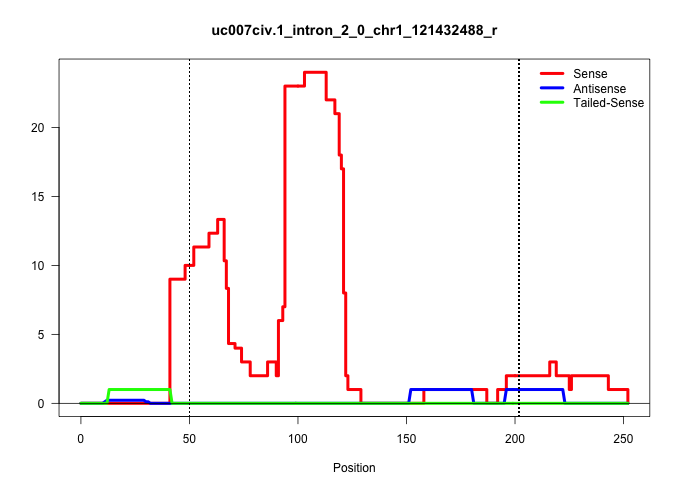
| Gene: AK084486 | ID: uc007civ.1_intron_2_0_chr1_121432488_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(1) TDRD1.ip |
(17) TESTES |
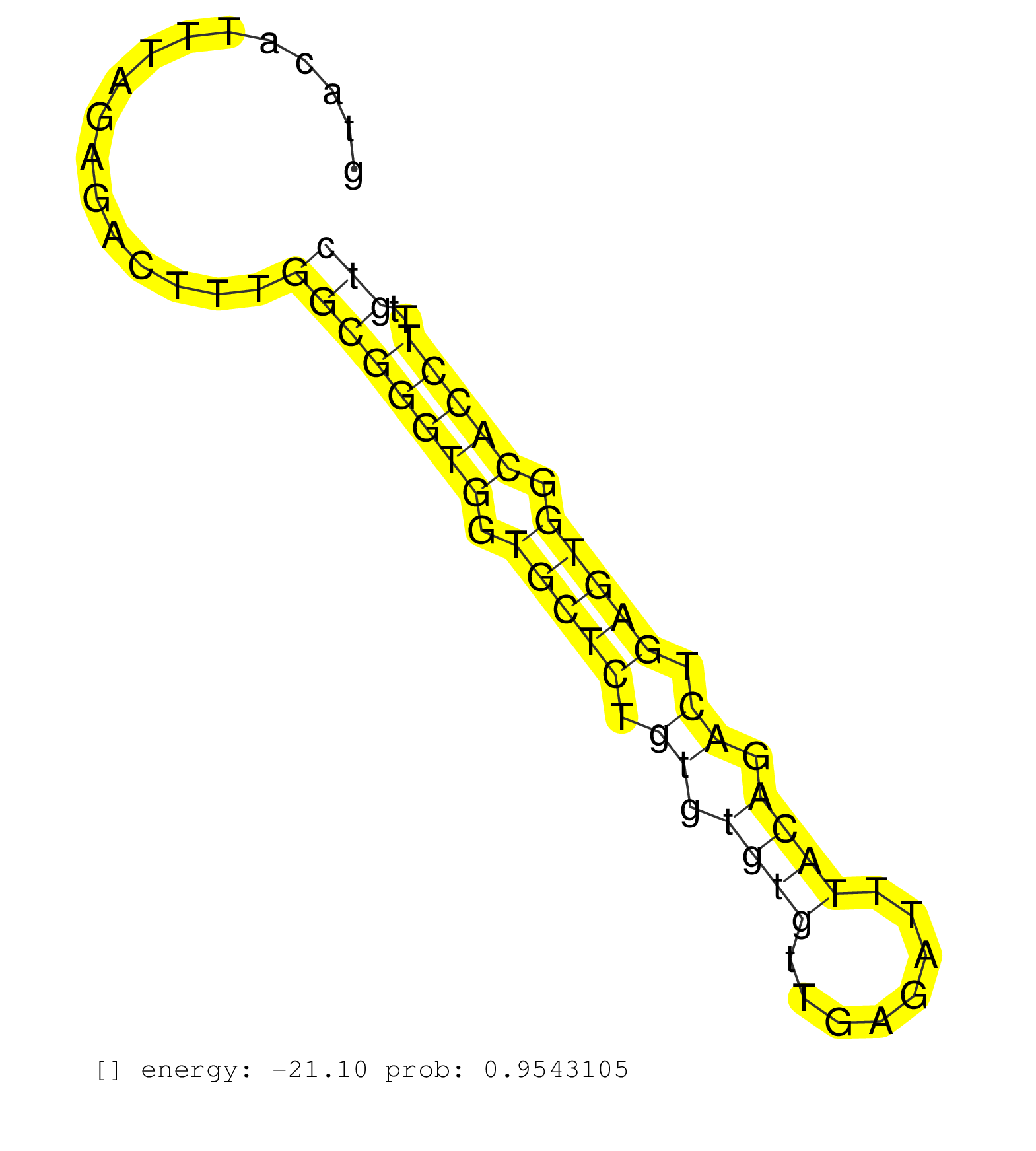
| TTAACAAGAAGAGTTAGCCAGAAGCAGAAGTACAGAAGCCATAGAGCAAGGTTGGTACATTTAGAGACTTTGGCGGGTGGTGCTCTGTGTGTGTTGAGATTTACAGACTGAGTGGCACCTTTGTCATGCAGCCAGCTGTGCTAAGGTCTGGTGCCTGTTACAGATATGCCTTAATTCTTTTACCTCCTCCTCTGCCTATCAGTCTTTGGTGCCTTTTAAATTATCATGTACACATGTGTGGGCTTGAGGTGT .......................................................................((((((((.(((((.((.((((........)))).)).))))).)))))..)))............................................................................................................................... ......................................................55....................................................................125............................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................TGAGATTTACAGACTGAGTGGCACCTT................................................................................................................................... | 27 | 1 | 9.00 | 9.00 | 2.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................TGAGATTTACAGACTGAGTGGCACCTTT.................................................................................................................................. | 28 | 1 | 6.00 | 6.00 | 3.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................TAGAGCAAGGTTGGTACATTTAGAGAC........................................................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................TGTTGAGATTTACAGACTGAGTGGCACC..................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TAGAGCAAGGTTGGTACATTTAGAG.......................................................................................................................................................................................... | 25 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................GTGTGTGTTGAGATTTACAGACTGAGT........................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................TAGAGCAAGGTTGGTACATTTAGAGA......................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ...........................................................TTTAGAGACTTTGGCGGGTGGTGCTCT...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................TTGAGATTTACAGACTGAGTGGCACCT.................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGGTACATTTAGAGACTTTGGCGGGT.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TGTACACATGTGTGGGCTTGAGGTGT | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TACAGATATGCCTTAATTCTTTTACCTCC................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................GAGACTTTGGCGGGTGGTGCTCTGTGT.................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGAGATTTACAGACTGAGTGGCACCTTTG................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................TGCCTATCAGTCTTTGGTGCCTTTTAA................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................AGGTTGGTACATTTAGAGACTTTGGC.................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TGTTGAGATTTACAGACTGAGTGGCA....................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............TTAGCCAGAAGCAGAAGTACAGAAGCCAg.................................................................................................................................................................................................................. | 29 | g | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TATCAGTCTTTGGTGCCTTTTAAATTATC........................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TGGTACATTTAGAGACTTTGGCGGG............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................................TAAATTATCATGTACACATGTGTGGGC......... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CAGACTGAGTGGCACCTTTGTCATGC........................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................TGGTACATTTAGAGACTTT..................................................................................................................................................................................... | 19 | 6 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | 0.17 |
| TTAACAAGAAGAGTTAGCCAGAAGCAGAAGTACAGAAGCCATAGAGCAAGGTTGGTACATTTAGAGACTTTGGCGGGTGGTGCTCTGTGTGTGTTGAGATTTACAGACTGAGTGGCACCTTTGTCATGCAGCCAGCTGTGCTAAGGTCTGGTGCCTGTTACAGATATGCCTTAATTCTTTTACCTCCTCCTCTGCCTATCAGTCTTTGGTGCCTTTTAAATTATCATGTACACATGTGTGGGCTTGAGGTGT .......................................................................((((((((.(((((.((.((((........)))).)).))))).)))))..)))............................................................................................................................... ......................................................55....................................................................125............................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................GCCTGTTACAGATATGCCTTAATTCTTTT....................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TATCAGTCTTTGGTGCCTTTTAAATTA............................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............GTTAGCCAGAAGCAGAAG.............................................................................................................................................................................................................................. | 18 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - |
| ...........AGTTAGCCAGAAGCAGAAGTA............................................................................................................................................................................................................................ | 21 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - |