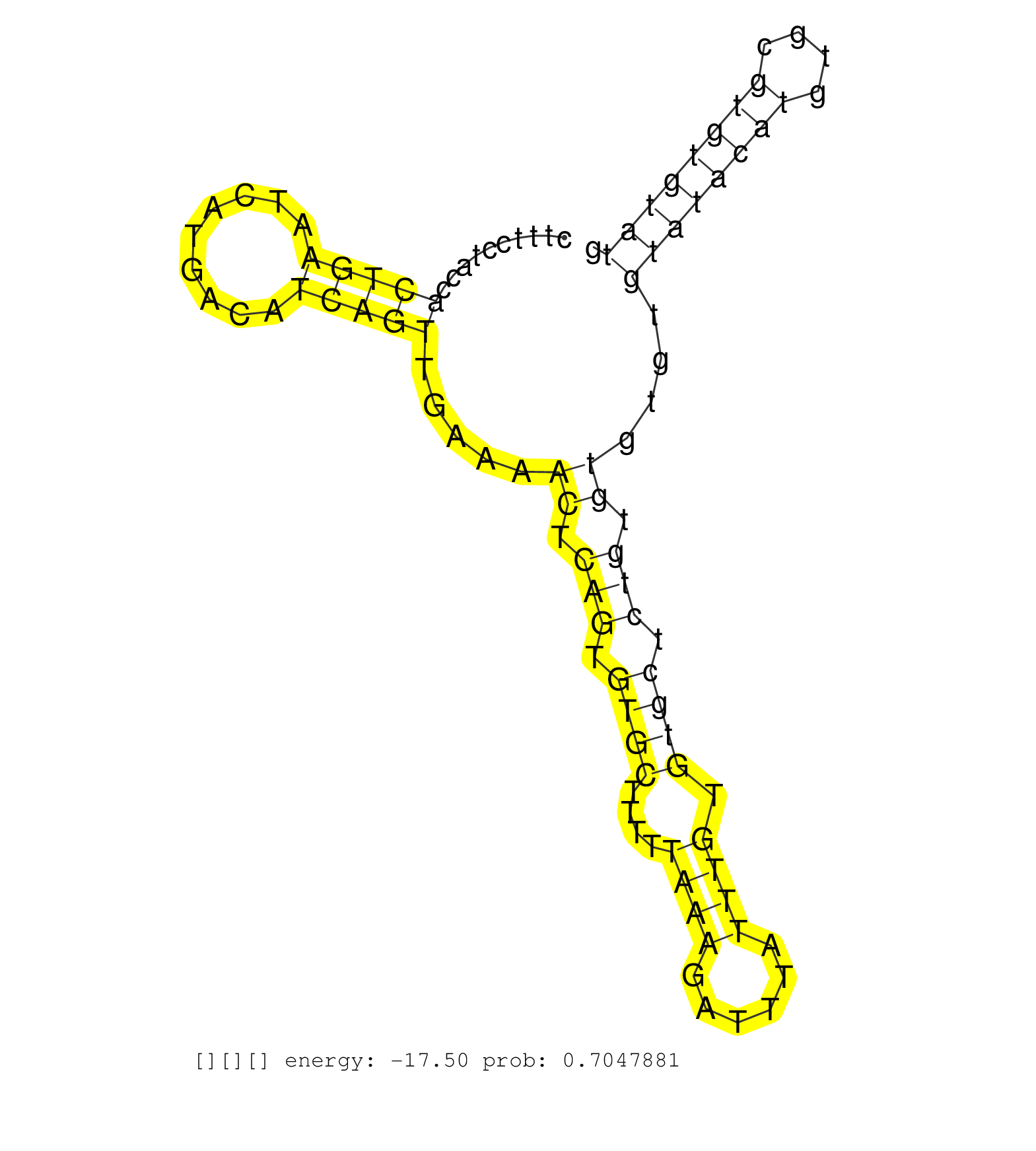
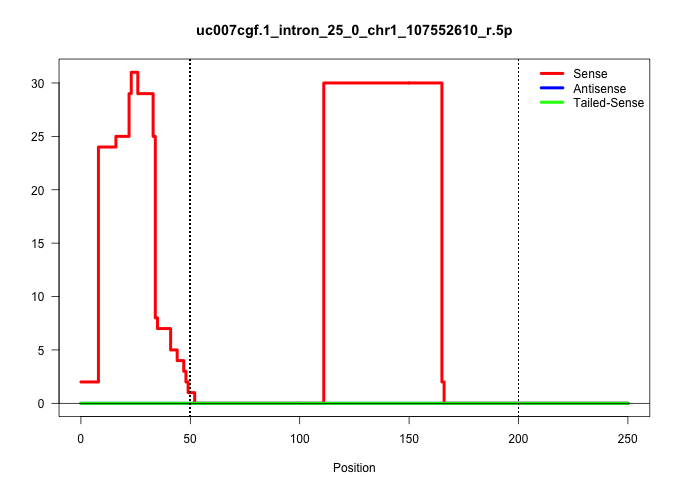
| Gene: Pign | ID: uc007cgf.1_intron_25_0_chr1_107552610_r.5p | SPECIES: mm9 |
 |
 |
|
(4) PIWI.ip |
(2) PIWI.mut |
(11) TESTES |
| TGTAGCCTTGATAGCTGGATTTTATGAGGATGTCAGTGCAGTTGCCAAAGGTTGTGATCTGAAATAATTTCATAAATAAAATGCTTGTCATATAAGCACTCTTTCCTACCACTGAATCATGACATCAGTTGAAAACTCAGTGTGCTTTTTAAAGATTTATTTGTGTGCTCTGTGTGTGTGTATACATGTGCGTGTGTATGCTTGTATGTTTGTGCACAATGTGAATGCAAGTGTTCATGGAGATCAGAAG ..............................................................................................................(((((.........))))).....((.(((.((((....((((......)))).)))).))).))....((((((((....))))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................CTGAATCATGACATCAGTTGAAAACTCAGTGTGCTTTTTAAAGATTTATTTG....................................................................................... | 52 | 1 | 30.00 | 30.00 | 30.00 | - | - | - | - | - | - | - | - | - | - |
| ........TGATAGCTGGATTTTATGAGGATGTC........................................................................................................................................................................................................................ | 26 | 1 | 17.00 | 17.00 | - | 7.00 | 1.00 | 3.00 | - | 2.00 | 2.00 | - | 1.00 | - | 1.00 |
| ........TGATAGCTGGATTTTATGAGGATGT......................................................................................................................................................................................................................... | 25 | 1 | 4.00 | 4.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - |
| ................GGATTTTATGAGGATGTCAGTGCAG................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................TATGAGGATGTCAGTGCAG................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................TATGAGGATGTCAGTGCAGTTGCCAA.......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................TATGAGGATGTCAGTGCAGTTGCCAAAGGT...................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................ATGAGGATGTCAGTGCAGTTG.............................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................TATGAGGATGTCAGTGCAGTTGCCAAA......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................ATGAGGATGTCAGTGCAGTTGCCA........................................................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........TGATAGCTGGATTTTATGAGGATGTCA....................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| TGTAGCCTTGATAGCTGGATTTTATGAGGATGTCAGTGCAGTTGCCAAAGGTTGTGATCTGAAATAATTTCATAAATAAAATGCTTGTCATATAAGCACTCTTTCCTACCACTGAATCATGACATCAGTTGAAAACTCAGTGTGCTTTTTAAAGATTTATTTGTGTGCTCTGTGTGTGTGTATACATGTGCGTGTGTATGCTTGTATGTTTGTGCACAATGTGAATGCAAGTGTTCATGGAGATCAGAAG ..............................................................................................................(((((.........))))).....((.(((.((((....((((......)))).)))).))).))....((((((((....))))))))................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|