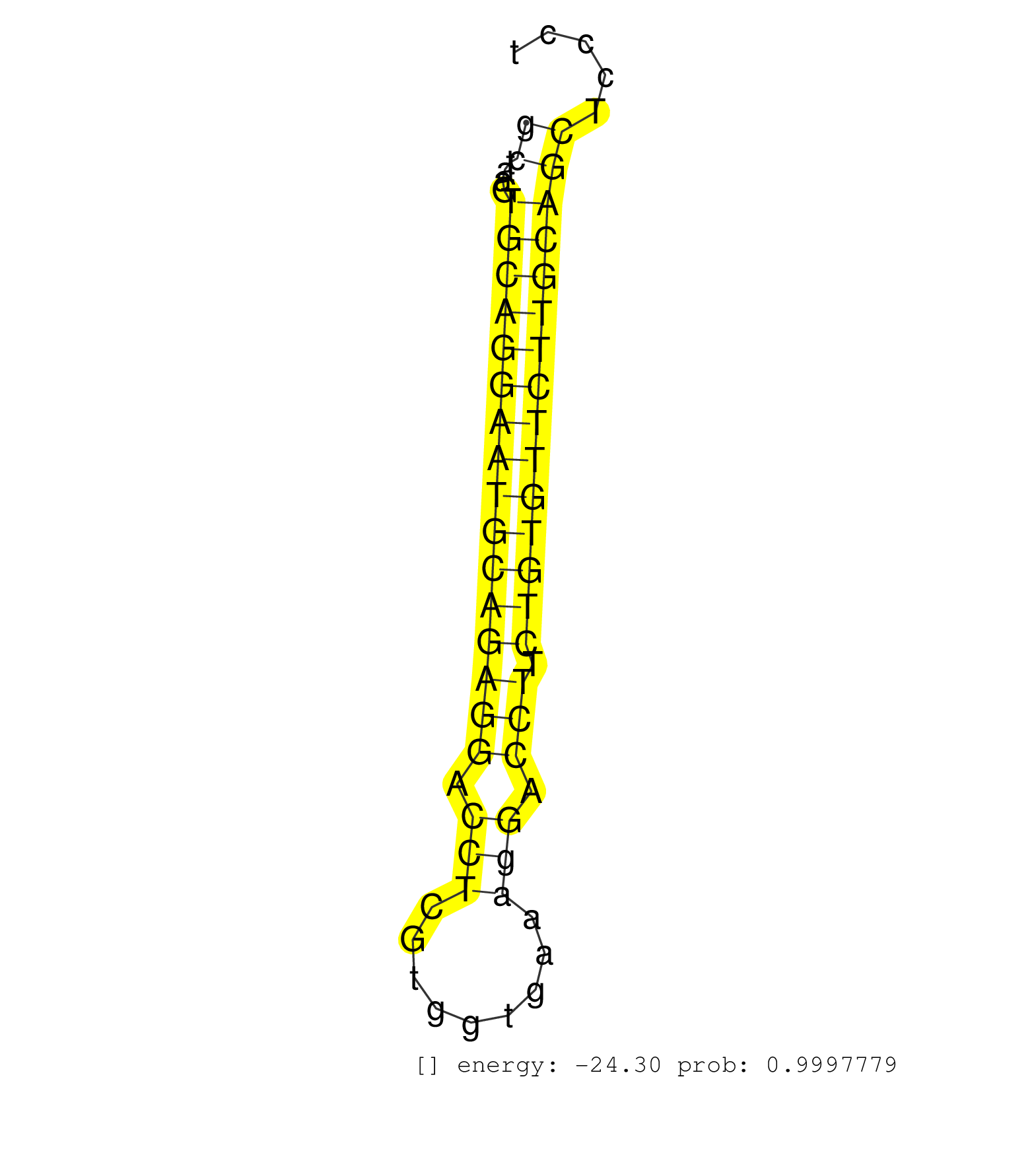

| Gene: Sned1 | ID: uc007cdm.1_intron_13_0_chr1_95171038_f | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(2) OVARY |
(1) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| ATACAAGTGTGACTGCCCTCCAGGATTCTCTGGACGGCACTGTGAGATAGGTAAGTAGGAAGCAGCCAGGGGTACTCGCCTCAGCACACATGAAAACTGGGTCAGTGGGCAGGACTGGGCTGGGAGGGGTCAGGTAAGCTAAGTGCAGGAATGCAGAGGACCTCGTGGTGAAAGGACCTTCTGTGTTCTTGCAGCTCCCTCACCCTGCTTCCGGAGCCCATGTATGAATGGGGGTACCTGTGAG .........................................................................................................................................((....((((((((((((((((.(((.........))).))).)))))))))))))))................................................. .........................................................................................................................................138...........................................................200.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................GACCTTCTGTGTTCTTGCAGtt................................................ | 22 | tt | 5.00 | 0.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................GTGCAGGAATGCAGAGGACCTat............................................................................... | 23 | at | 3.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................GACCTTCTGTGTTCTTGCAGttt............................................... | 23 | ttt | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................AGTGCAGGAATGCAGAGGACCTttaa............................................................................. | 26 | ttaa | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGTGCAGGAATGCAGAGGACCTtt............................................................................... | 25 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................GTGCAGGAATGCAGAGGACCTta............................................................................... | 23 | ta | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGTGCAGGAATGCAGAGGACCTat............................................................................... | 24 | at | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................AGGATTCTCTGGACGGCACTGTGAGATA................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CAGGAATGCAGAGGACCTttat............................................................................. | 22 | ttat | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GTGCAGGAATGCAGAGGACCTaa............................................................................... | 23 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................ATTCTCTGGACGGCACTGTG........................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................TCCAGGATTCTCTGGACGGCACTGTGAG...................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GTGCAGGAATGCAGAGGACCTaat.............................................................................. | 24 | aat | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGTGCAGGAATGCAGAGGACCT................................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTGTGTTCTTGCAGttt............................................... | 18 | ttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ACAAGTGTGACTGCCCTCC............................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGTGCAGGAATGCAGAGGACagt................................................................................ | 24 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................AGTGCAGGAATGCAGAGGACCTa................................................................................ | 23 | a | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGTGCAGGAATGCAGAGGACCTaa............................................................................... | 24 | aa | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................GGATTCTCTGGACGGCACTGTGAGA..................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GTGCAGGAATGCAGAGGACCT................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GTGCAGGAATGCAGAGGACCTtaa.............................................................................. | 24 | taa | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................GTGCAGGAATGCAGAGGACCTtt............................................................................... | 23 | tt | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ATACAAGTGTGACTGCCCTCCAGGATTCTCTGGACGGCACTGTGAGATAGGTAAGTAGGAAGCAGCCAGGGGTACTCGCCTCAGCACACATGAAAACTGGGTCAGTGGGCAGGACTGGGCTGGGAGGGGTCAGGTAAGCTAAGTGCAGGAATGCAGAGGACCTCGTGGTGAAAGGACCTTCTGTGTTCTTGCAGCTCCCTCACCCTGCTTCCGGAGCCCATGTATGAATGGGGGTACCTGTGAG .........................................................................................................................................((....((((((((((((((((.(((.........))).))).)))))))))))))))................................................. .........................................................................................................................................138...........................................................200.......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................AGGACCTTCTGTGTcagt.......................................................... | 18 | cagt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |