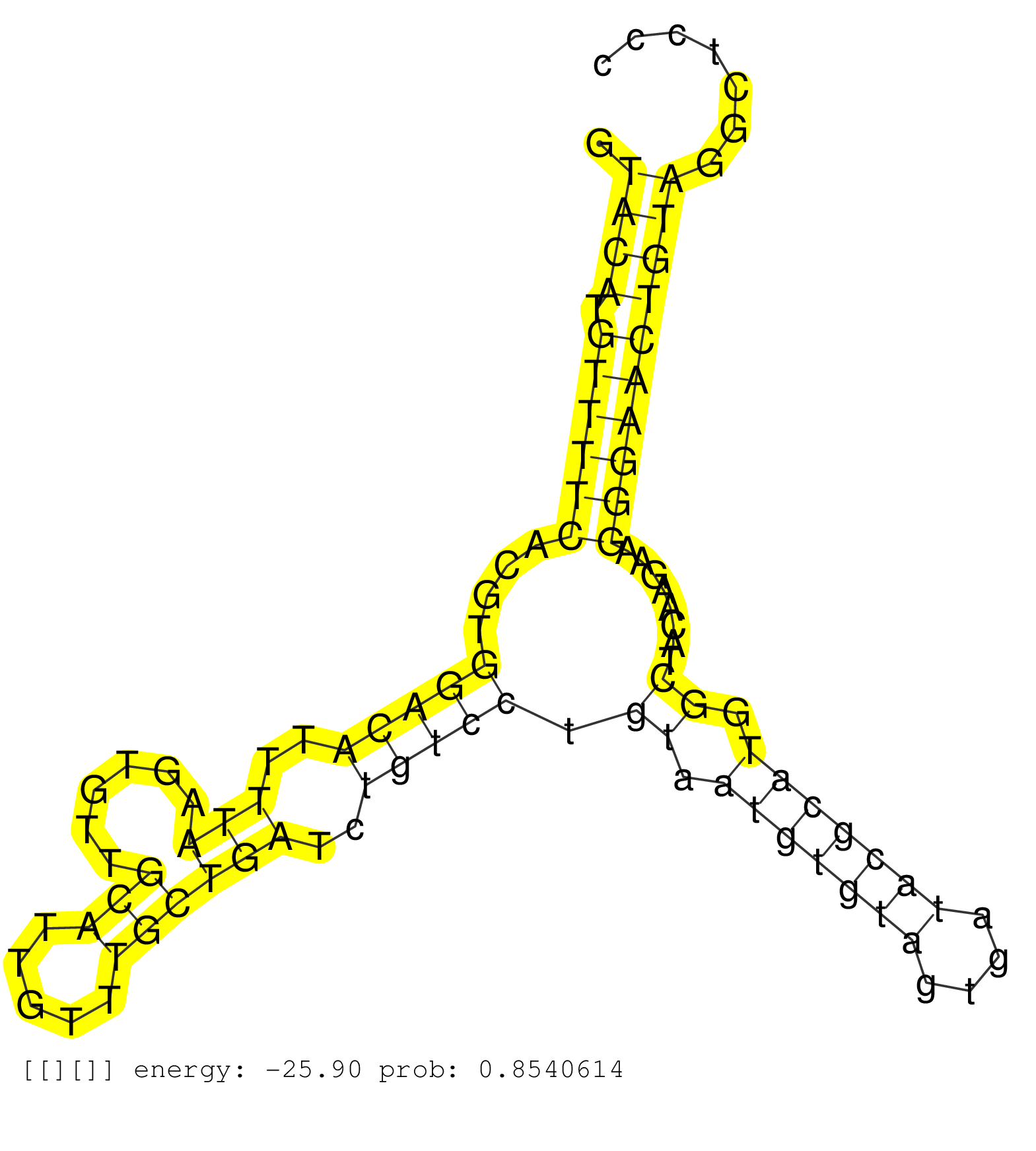
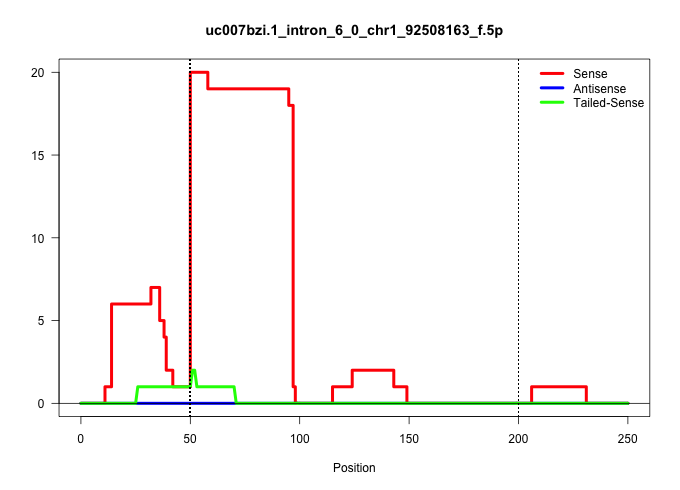
| Gene: Cops8 | ID: uc007bzi.1_intron_6_0_chr1_92508163_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(15) TESTES |
| AGCCTCAGGAACCCTGGATGTCTCCTTGAACAGGTTTATCCCCTTATCAGGTACATGTTTTCACGTGGACATTTTAAGTGTTGCATTGTTTGCTGATCTGTCCTGTAATGTGTAGTGATACGCATGGCTACAAGAAGGGAACTGTAGGCTCCCCTGCAGCTTAGCTGGAGTGGTGGCTCCTGATGTCCAGCCACAGAGCCTGCCTATCCTCCTTTGAGAGTCTCTGGGTTCTGGACTGTATTCTGAGGGC ...................................................((((.((((((....(((((..(((......(((.....))))))..))))).((.(((((((....))))))).))........))))))))))........................................................................................................ ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTACATGTTTTCACGTGGACATTTTAAGTGTTGCATTGTTTGCTGATCTGTC.................................................................................................................................................... | 52 | 1 | 16.00 | 16.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GAAGGGAACTGTAGGCTCCCCTGCAGC.......................................................................................... | 27 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGGATGTCTCCTTGAACAGGTTTAT................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGGATGTCTCCTTGAACAGGTT...................................................................................................................................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............TGGATGTCTCCTTGAACAGGTTTATCCC................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................TACATGTTTTCACGTGtgtc................................................................................................................................................................................... | 20 | tgtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................TGGCTACAAGAAGGGAACTGTAGGC..................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................GGTTTATCCCCTTATCAGGTACATGT................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TGAACAGGTTTATCCCCTTATCAGagc..................................................................................................................................................................................................... | 27 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........CCCTGGATGTCTCCTTGAACAGGTTTA.................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GTTCTGGACTGTATTCTG..... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TCCTCCTTTGAGAGTCTCTGGGTTC................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................ACTGTAGGCTCCCCTGC............................................................................................. | 17 | 2 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............CTGGATGTCTCCTTGAACAGG........................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................TGATACGCATGGCTACAAGAAGGGAACT........................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................AGAAGGGAACTGTAG....................................................................................................... | 15 | 14 | 0.07 | 0.07 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.07 |
| AGCCTCAGGAACCCTGGATGTCTCCTTGAACAGGTTTATCCCCTTATCAGGTACATGTTTTCACGTGGACATTTTAAGTGTTGCATTGTTTGCTGATCTGTCCTGTAATGTGTAGTGATACGCATGGCTACAAGAAGGGAACTGTAGGCTCCCCTGCAGCTTAGCTGGAGTGGTGGCTCCTGATGTCCAGCCACAGAGCCTGCCTATCCTCCTTTGAGAGTCTCTGGGTTCTGGACTGTATTCTGAGGGC ...................................................((((.((((((....(((((..(((......(((.....))))))..))))).((.(((((((....))))))).))........))))))))))........................................................................................................ ..................................................51....................................................................................................153............................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT3() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................................................CCAGCCACAGAGCCTggg................................................. | 18 | ggg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |