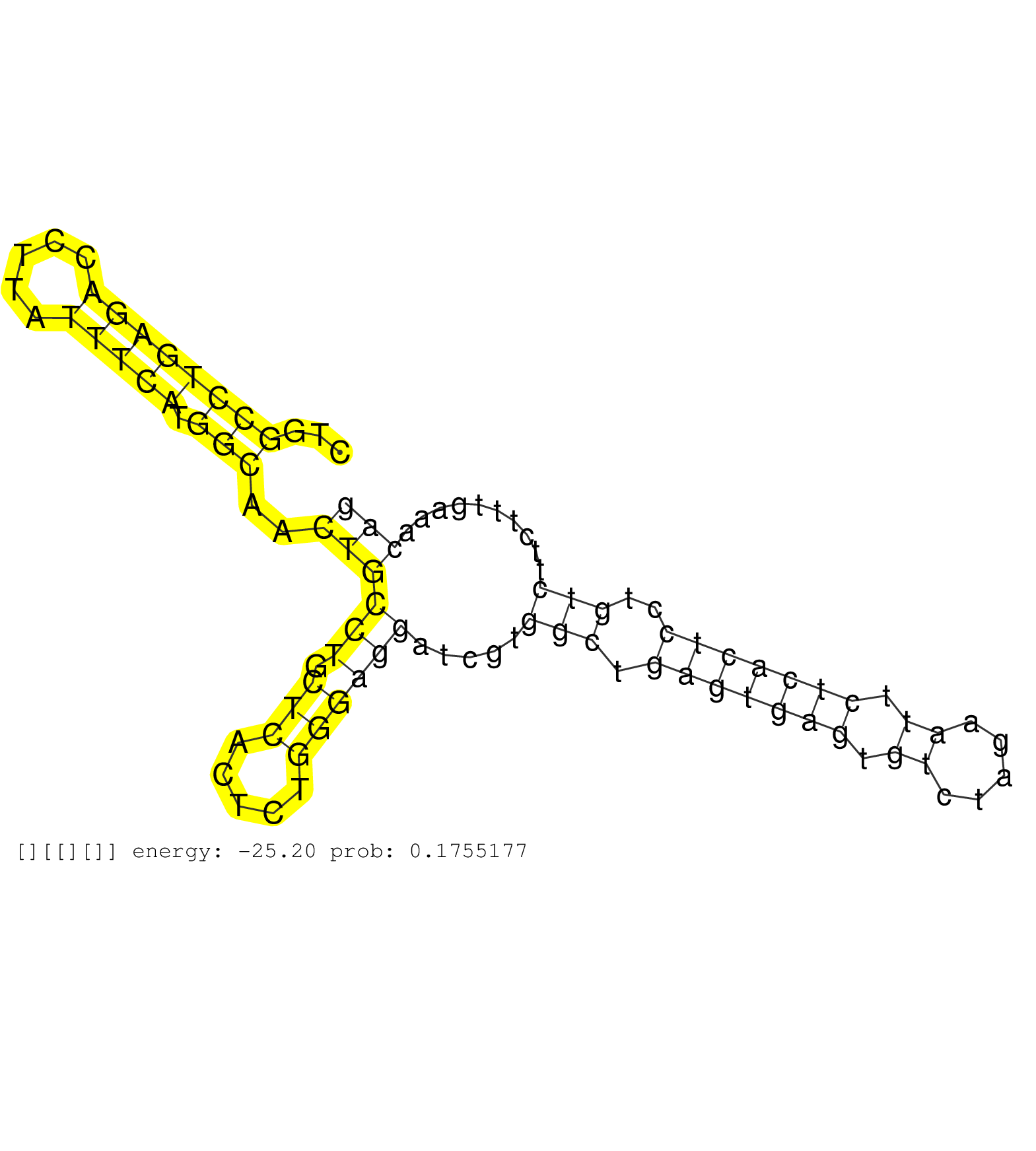

| Gene: Armc9 | ID: uc007bvd.1_intron_18_0_chr1_88109681_f.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| TCAAATATCCTGTAATAGTTGTCAGCTGTGCCCCCCTCACTTTTTTAGAGGGATTTTGAGCAGGGGATAGGTGGCTTCTGAGACCTGGTGGGACTTTATGCTGGCCTGAGACCTTATTTCATGGCAACTGCCTGCTCACTCTGGGAGGATCGTGGCTGAGTGAGTGTCTAGAATTCTCACTCCTGTCTTCTTTGAAACAGGAGGATCATGACATCATGGAAGCTGACCTGGACAAAGATGAACTGATCCA .......................................................................................................((((((((.....))))).)))..((((((.(((.....)))))).....(((.(((((((.((.....)).)))))))..)))..........))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................GGACTTTATGCTGGCCTGAGACCTTATTTCATGGCAACTGCCTGCTCACTCT............................................................................................................ | 52 | 1 | 23.00 | 23.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGGAGGATCGTGGCTGAGTGAGT..................................................................................... | 24 | 1 | 7.00 | 7.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAAGCTGACCTGGACAAAGATGAAC....... | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................................TGGAAGCTGACCTGGACAAAGATGAA........ | 26 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................GCTGACCTGGACAAAGATGAACTGATC.. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TGACATCATGGAAGCTGACCTGGA.................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................TGACATCATGGAAGCTGACCTGGAC................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................CCCCCTCACTTTTTagtc......................................................................................................................................................................................................... | 18 | agtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................................................................................................................CATGACATCATGGAAGCTGACCTGGA.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TGACATCATGGAAGCTGACCTG.................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................TGACATCATGGAAGCTGACCTGtact................ | 26 | tact | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TCATGGAAGCTGACCTGGACAAAGATG.......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TGACATCATGGAAGCTGACCTGG................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................................TGACATCATGGAAGCTGACCTGGACA................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| TCAAATATCCTGTAATAGTTGTCAGCTGTGCCCCCCTCACTTTTTTAGAGGGATTTTGAGCAGGGGATAGGTGGCTTCTGAGACCTGGTGGGACTTTATGCTGGCCTGAGACCTTATTTCATGGCAACTGCCTGCTCACTCTGGGAGGATCGTGGCTGAGTGAGTGTCTAGAATTCTCACTCCTGTCTTCTTTGAAACAGGAGGATCATGACATCATGGAAGCTGACCTGGACAAAGATGAACTGATCCA .......................................................................................................((((((((.....))))).)))..((((((.(((.....)))))).....(((.(((((((.((.....)).)))))))..)))..........))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|