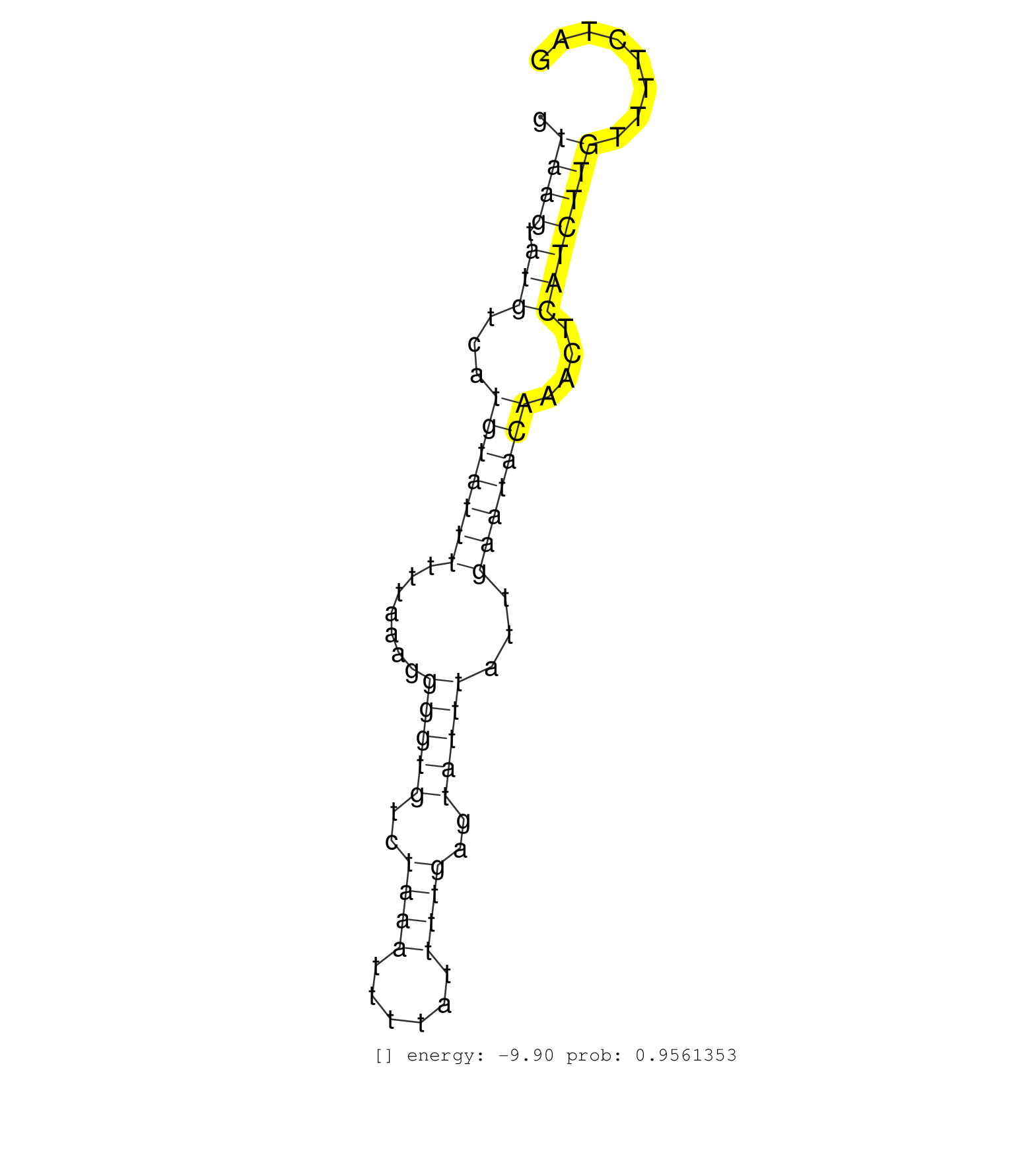
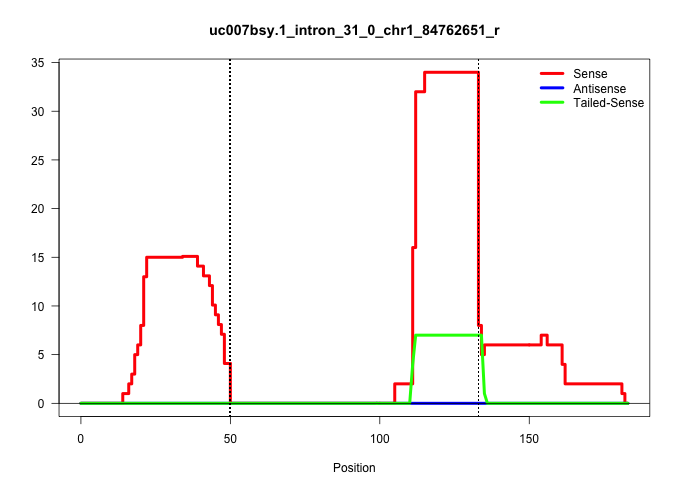
| Gene: Trip12 | ID: uc007bsy.1_intron_31_0_chr1_84762651_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(6) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(23) TESTES |
| TATGTTTTTAACAGATAACATTACTGCAGATGGAACATAATTTTGACATTGTAAGTATGTCATGTATTTTTTAAAGGGGTGTCTAAATTTTATTTTGAGTATTTATTGAATACAAACTCATCTTGTTTTCTAGATGAATCATGCTTGCCGAGCCTTAACATATATGATGGAAGCTCTCCCCCG ...................................................((((.(((...(((((((.......(((((..((((......))))..)))))...)))))))....))))))).......................................................... ..................................................51................................................................................133................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................CAAACTCATCTTGTTTTCTAG.................................................. | 21 | 1 | 13.00 | 13.00 | 4.00 | 2.00 | 2.00 | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................ACAAACTCATCTTGTTTTCTAG.................................................. | 22 | 1 | 12.00 | 12.00 | 4.00 | 4.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................TACTGCAGATGGAACATAATTTTGACATT..................................................................................................................................... | 29 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................CAAACTCATCTTGTTTTCTAGA................................................. | 22 | 1 | 4.00 | 4.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ACAAACTCATCTTGTTTTCTAGA................................................. | 23 | 1 | 4.00 | 4.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGAATCATGCTTGCCGAGCCTTAACAT...................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................ACAAACTCATCTTGTTTTCTAGAa................................................ | 24 | a | 3.00 | 4.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................TACTGCAGATGGAACATAATTTTGACA....................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................ACTCATCTTGTTTTCTAGA................................................. | 19 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................CAAACTCATCTTGTTTTCTAGAa................................................ | 23 | a | 2.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................TTGAATACAAACTCATCTTGTTTTCTAG.................................................. | 28 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGAATCATGCTTGCCGAGCCTTAACATA..................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ACAAACTCATCTTGTTTTCTAGt................................................. | 23 | t | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CAAACTCATCTTGTTTTCTAtaa................................................ | 23 | taa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................ACTGCAGATGGAACATAATTTTGA......................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................ATTACTGCAGATGGAACATAATTT............................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTAACATATATGATGGAAGCTCTCCCC.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................ACATTACTGCAGATGGAACATAATTTTG.......................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................AACATTACTGCAGATGGAACATAATTTT........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TTACTGCAGATGGAACATAATTTTGAC........................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TTACTGCAGATGGAACATAAT.............................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................................................................................GAATCATGCTTGCCGAGCCTT........................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................CATTACTGCAGATGGAACATAATTTTGACA....................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................ACAAACTCATCTTGTTTTCTAGAaa............................................... | 25 | aa | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............ATAACATTACTGCAGATGGAACATA................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ATATGATGGAAGCTCTCCCCC. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................ACTGCAGATGGAACATAATTTTGACATT..................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................CATTACTGCAGATGGAACATAATTTT........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................ACATAATTTTGACATT..................................................................................................................................... | 16 | 11 | 0.09 | 0.09 | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TATGTTTTTAACAGATAACATTACTGCAGATGGAACATAATTTTGACATTGTAAGTATGTCATGTATTTTTTAAAGGGGTGTCTAAATTTTATTTTGAGTATTTATTGAATACAAACTCATCTTGTTTTCTAGATGAATCATGCTTGCCGAGCCTTAACATATATGATGGAAGCTCTCCCCCG ...................................................((((.(((...(((((((.......(((((..((((......))))..)))))...)))))))....))))))).......................................................... ..................................................51................................................................................133................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) |
|---|