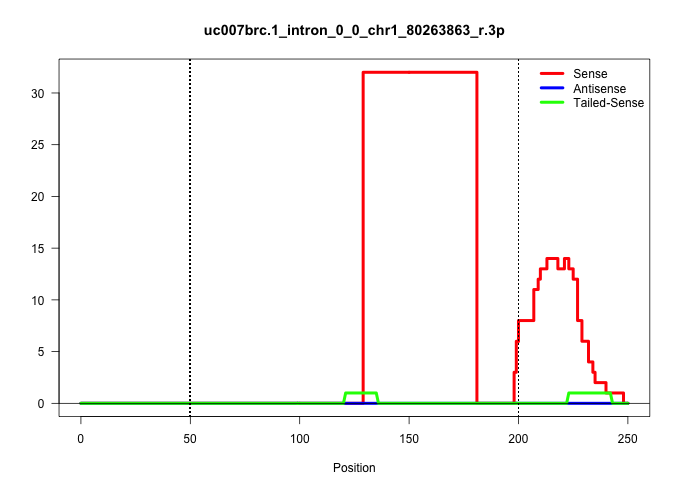
| Gene: Cul3 | ID: uc007brc.1_intron_0_0_chr1_80263863_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(13) TESTES |
| TTCTTGTCTAGTTCTTACCTCACATCCCCAAGTACTAAGATTATAGGTATTAGGCACCATGCTTGGTGCATCTAACTTTTTTTAAATCAGGTCACTAAATTTTTCAGCTTTAAATGGCATGTCGGCTCTAGTAAAAAGATAGGGATTTTTGTTTGTCTATGTGGAAGTGTTTTAGTTAAGACTTTACTTTTTCTTTTTAGGTAACTCAGCAACTGAAGGCTCGATTCTTACCAAGTCCAGTTGTTATTAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................AGTAAAAAGATAGGGATTTTTGTTTGTCTATGTGGAAGTGTTTTAGTTAAGA..................................................................... | 52 | 1 | 32.00 | 32.00 | 32.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGTAACTCAGCAACTGAAGGCTCGATTC....................... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGCAACTGAAGGCTCGATTCTTACC.................. | 25 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................TCGGCTCTAGTggtt.................................................................................................................. | 15 | ggtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................CAACTGAAGGCTCGATTCTTACCAAG............... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................AACTGAAGGCTCGATTCTT..................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................AGCAACTGAAGGCTCGATTCTTACCAA................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTAACTCAGCAACTGAAGGCTCG........................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................GTAACTCAGCAACTGAAGGCTCGATTC....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................ATTCTTACCAAGTCCAGTTa....... | 20 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTAACTCAGCAACTGAAGGCTCGAT......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................................CGATTCTTACCAAGTCCAGTTGTTATT.. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGAAGGCTCGATTCTTACCAAGTCCAG.......... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................GTAACTCAGCAACTGAAG................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTAACTCAGCAACTGAAGGCTCGATTCTT..................... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| TTCTTGTCTAGTTCTTACCTCACATCCCCAAGTACTAAGATTATAGGTATTAGGCACCATGCTTGGTGCATCTAACTTTTTTTAAATCAGGTCACTAAATTTTTCAGCTTTAAATGGCATGTCGGCTCTAGTAAAAAGATAGGGATTTTTGTTTGTCTATGTGGAAGTGTTTTAGTTAAGACTTTACTTTTTCTTTTTAGGTAACTCAGCAACTGAAGGCTCGATTCTTACCAAGTCCAGTTGTTATTAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|