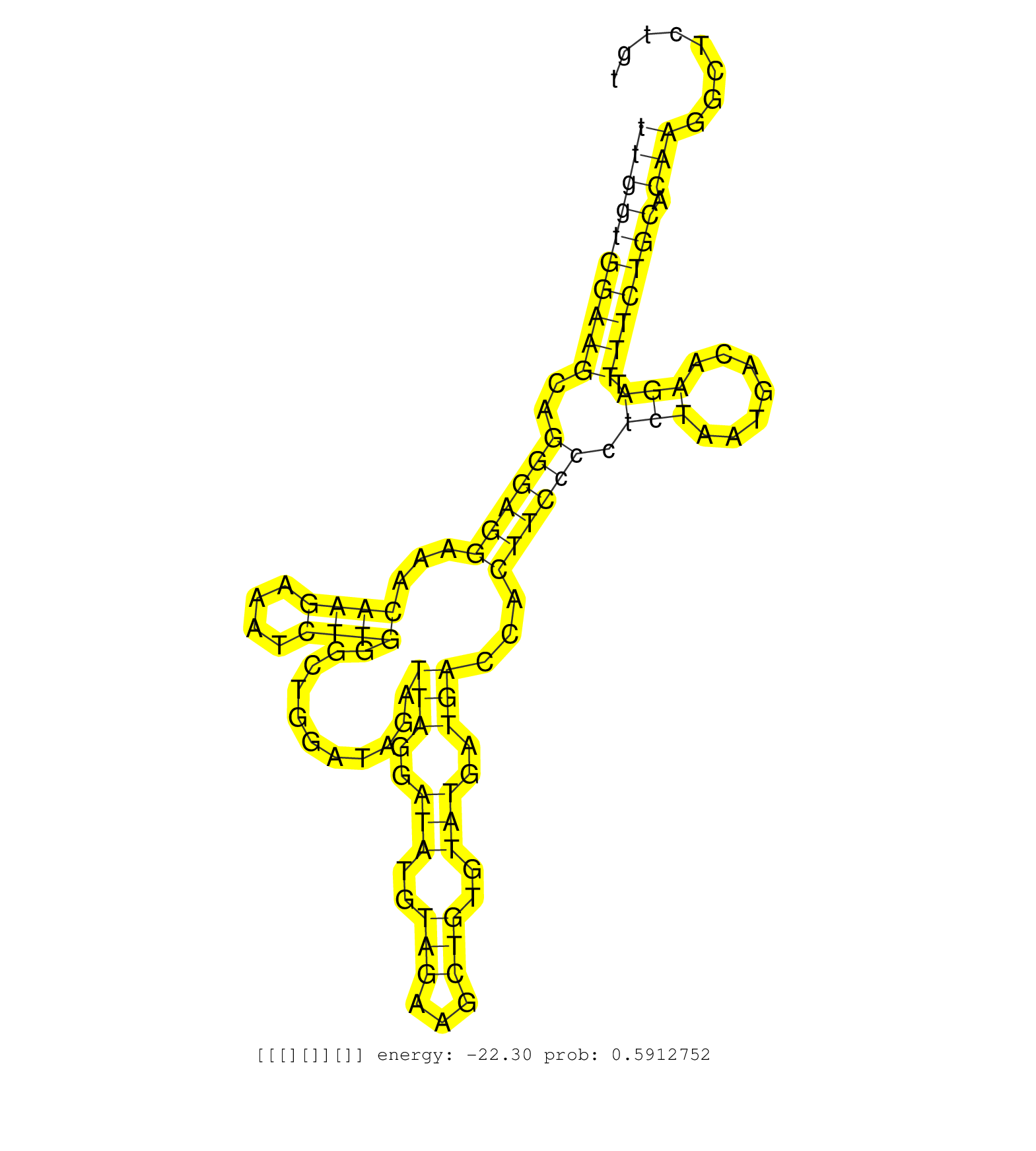
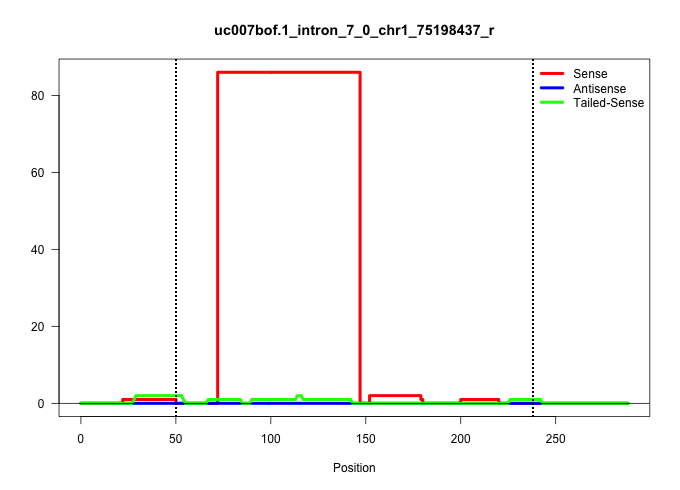
| Gene: Glb1l | ID: uc007bof.1_intron_7_0_chr1_75198437_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(13) TESTES |
| CCACTTTAGGTATATGTTCCATGGAGGTACCAACTTTGGATACTGGAATGGTGAGTATCAATTCCCTTTGGTGGAAGCAGGGAGGAAACAAGAAATCTTGGGCTGGATAGATTAGGATATGTAGAAGCTGTGTATGATGACCACTTCCCCTCTAATGACAAGATTTTCTGCACAAGGCTCTGTGCTTAGTGCGCATCAGCTAAGATGATGTTGAATTAATTAAATTACTTTAACCCAGGAGCTGACGAGAAGGGACGCTTCCTTCCAATTACTACCAGCTACGACTAT ...................................................................((((((((((..((((((...((((....))))...........(((..(((..(((...)))..)))..)))...)))))).(((.......))).))))))).)))................................................................................................................. ...................................................................68.................................................................................................................183....................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................GGAAGCAGGGAGGAAACAAGAAATCTTGGGCTGGATAGATTAGGATATGTAG.................................................................................................................................................................... | 52 | 1 | 91.00 | 91.00 | 91.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TACCAACTTTGGATACTGGAATGGagct......................................................................................................................................................................................................................................... | 28 | agct | 6.00 | 0.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TAATGACAAGATTTTCTGCACAAGGCT............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................GGAGGTACCAACTTTGGATACTGGAATG.............................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................................................................ACTTTAACCCAGGAGgt............................................. | 17 | gt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................CCAACTTTGGATACTGGAATGGagct......................................................................................................................................................................................................................................... | 26 | agct | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................................TAAGATGATGTTGAATTAAT.................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................................................................................................................GCTGACGAGAAGGGACGCTTC........................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................GGATATGTAGAAGCTGTGTATGATGACCt................................................................................................................................................. | 29 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................TTGGTGGAAGCAGGttca........................................................................................................................................................................................................... | 18 | ttca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................AGAAATCTTGGGCTGGATAGATTAGGt........................................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................ACCAACTTTGGATACTGGAATGGagc.......................................................................................................................................................................................................................................... | 26 | agc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................CTTTAACCCAGGAGCTGA........................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................................................................................TAATGACAAGATTTTCTGCACAAGGCTC............................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| CCACTTTAGGTATATGTTCCATGGAGGTACCAACTTTGGATACTGGAATGGTGAGTATCAATTCCCTTTGGTGGAAGCAGGGAGGAAACAAGAAATCTTGGGCTGGATAGATTAGGATATGTAGAAGCTGTGTATGATGACCACTTCCCCTCTAATGACAAGATTTTCTGCACAAGGCTCTGTGCTTAGTGCGCATCAGCTAAGATGATGTTGAATTAATTAAATTACTTTAACCCAGGAGCTGACGAGAAGGGACGCTTCCTTCCAATTACTACCAGCTACGACTAT ...................................................................((((((((((..((((((...((((....))))...........(((..(((..(((...)))..)))..)))...)))))).(((.......))).))))))).)))................................................................................................................. ...................................................................68.................................................................................................................183....................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................................AATTACTTTAACCCAgggt................................................... | 19 | gggt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |