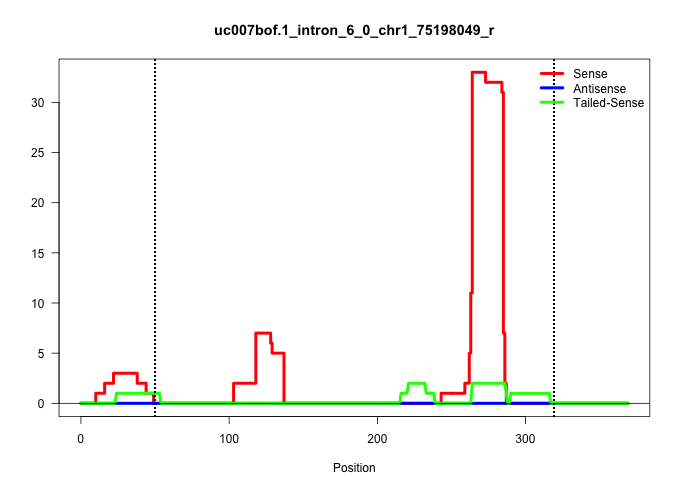
| Gene: Glb1l | ID: uc007bof.1_intron_6_0_chr1_75198049_r | SPECIES: mm9 |
 |
 |
|
(8) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(26) TESTES |
| CAGGGGACCCTACACCTAAGCTTTTTGCAATTCGAAATGTCATCAGCAAGGTGCCCTTTTATTAATTGGGAGAAGGGAGTGGGCCCATATTGTACAGGTGCCCTACAAACTGGTAGTGTTGTCTGGGGAGCAGAGCCCACTTAGCATTCCATGAAGCAATTCCCATTCATATAGAACCATCTGGGGAATTCCACATGTGAAGAATTATGGGTCTTCATGGCTCGCTGCAGCCCATCCCTGGGTTCCAGGTCAAGAACCTTCTTTTAGAGCCTTGGATTGTCTTGGTTTTCTTATGACTACTGAGAGAATTTGGGCTCAGTTCCAGGAAATTCCCTTGGGACCTTTACCTCCTCCCAGCCCCAAGATGAA ..................................................................................................................(((((((..((((........))))..)))))))(((.((.....(((((((((.....))).....))))))..((((...(((((((........)))))))(((((.(((.((((((....)))))).)))...((((....))))...))))).))))...)).))).................................................................................... .................................................................................................................114............................................................................................................................................................................289.............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................................................................................................................TAGAGCCTTGGATTGTCTTGG.................................................................................... | 21 | 1 | 16.00 | 16.00 | 6.00 | 4.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................TTAGAGCCTTGGATTGTCTTGG.................................................................................... | 22 | 1 | 7.00 | 7.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................TAGAGCCTTGGATTGTCTTGGT................................................................................... | 22 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................................................................TGAGAGAATTTGGGCTCAGa................................................. | 20 | a | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................................................................TAGAGCCTTGGATTGTCTTGGTa.................................................................................. | 23 | a | 2.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................TAGAGCCTTGGATTGTCTTGGTT.................................................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................TTTAGAGCCTTGGATTGTCTTGG.................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TACACCTAAGCTTTTTGCAATTCGAAAT........................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................................TCGCTGCAGCCCATCggt.................................................................................................................................. | 18 | ggt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................TAGAGCCTTGGATTGTCTTG..................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TAAGCTTTTTGCAATTCGAAATGTCATC..................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................TCTTATGACTACTGAGAGAATTTGGGCTC.................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................................................................................................................................................ATGGCTCGCTGCAacct........................................................................................................................................ | 17 | acct | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................TAGAGCCTTGGATTGTCTTGGTat................................................................................. | 24 | at | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................................................................TCTTTTAGAGCCTTGGATTGTCTTGGT................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................TACAAACTGGTAGTGTTGTCTGGGG................................................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TTGCAATTCGAAATGTCATCAGCAAGGacc........................................................................................................................................................................................................................................................................................................................... | 30 | acc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................TACAAACTGGTAGTGTTGTCTGGGGA................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TTTTGCAATTCGAAATGTCATCAGCAA................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................TTATGACTACTGAGAGAATTTGGGCTt.................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................TTTAGAGCCTTGGATTGTCTTGGT................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................TCCAGGTCAAGAACCTTCTTTTAGAGCCTT................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CAGGGGACCCTACACCTAAGCTTTTTGCAATTCGAAATGTCATCAGCAAGGTGCCCTTTTATTAATTGGGAGAAGGGAGTGGGCCCATATTGTACAGGTGCCCTACAAACTGGTAGTGTTGTCTGGGGAGCAGAGCCCACTTAGCATTCCATGAAGCAATTCCCATTCATATAGAACCATCTGGGGAATTCCACATGTGAAGAATTATGGGTCTTCATGGCTCGCTGCAGCCCATCCCTGGGTTCCAGGTCAAGAACCTTCTTTTAGAGCCTTGGATTGTCTTGGTTTTCTTATGACTACTGAGAGAATTTGGGCTCAGTTCCAGGAAATTCCCTTGGGACCTTTACCTCCTCCCAGCCCCAAGATGAA ..................................................................................................................(((((((..((((........))))..)))))))(((.((.....(((((((((.....))).....))))))..((((...(((((((........)))))))(((((.(((.((((((....)))))).)))...((((....))))...))))).))))...)).))).................................................................................... .................................................................................................................114............................................................................................................................................................................289.............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................................................................................TTTTCTTATGACTACaa..................................................................... | 17 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |