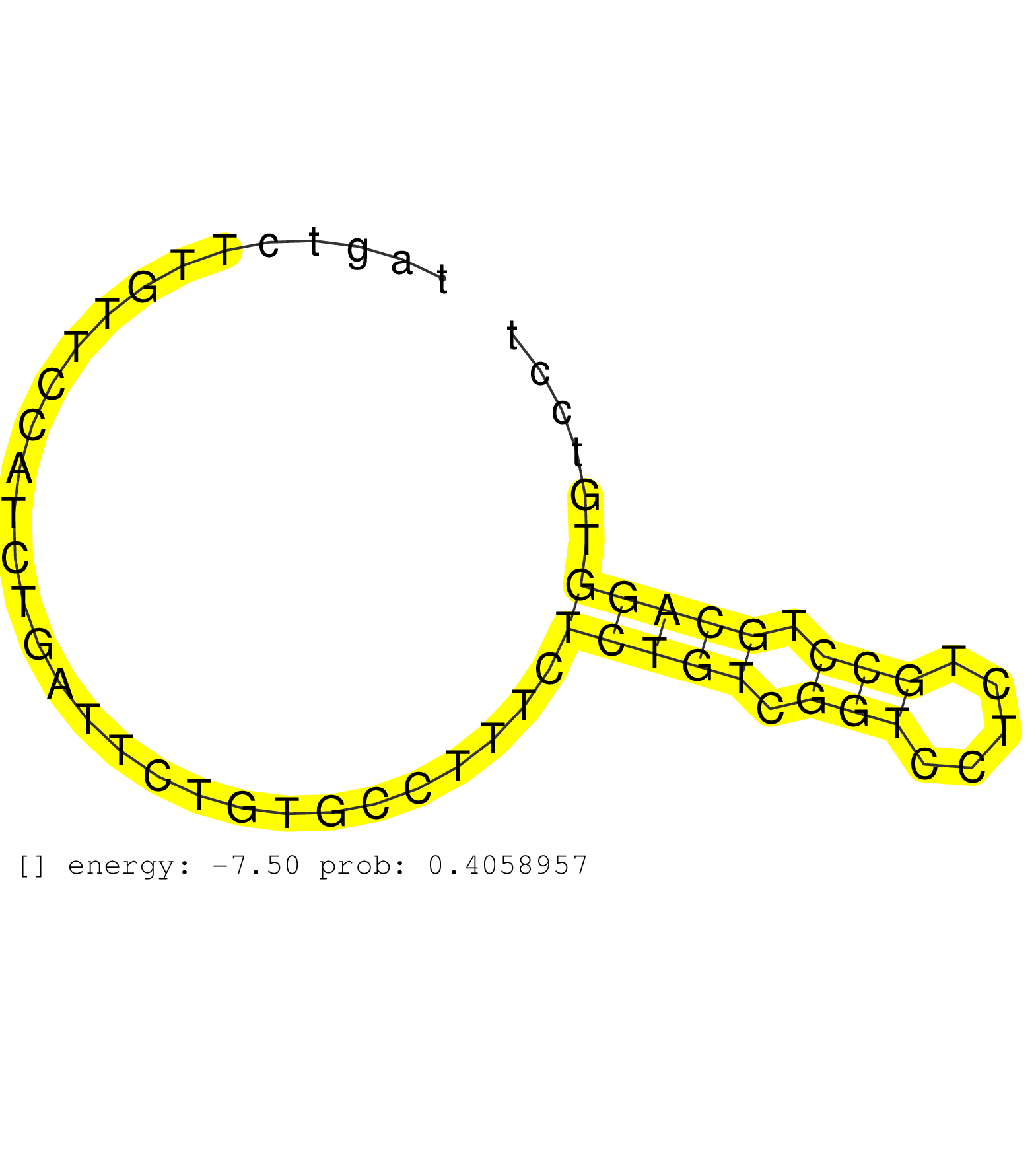

| Gene: Zfp142 | ID: uc007bmn.1_intron_6_0_chr1_74626502_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(14) TESTES |
| AAGACATATGCCAGATACCAGACAGGTTATCCCTGACCTATAGGCTTATTATAGCCCAGTACAGGGCCAGACTTGAAGTCTGTCTCTACTCCATGACTATGAAAAGATTCACCTACATTTCTTCTAGTGACTAACTCCTTCAAGTCTTAGTCTTGTTCCATCTGATTCTGTGCCTTTCTCTGTCGGTCCTCTGCCTGCAGGTGTCCTGGTAAAGGTGGTGGAGGTATACTTCTGTGAGCGCTGTGAACAG ..................................................................................................................................................................................(((((.(((.....))).)))))................................................. ...................................................................................................................................................148........................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................TTGTTCCATCTGATTCTGTGCCTTTCT....................................................................... | 27 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CTCTGTCGGTCCTCTGCCTGCAGGTG............................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........TGCCAGATACCAGACAGGTTATCCCTGAC..................................................................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............TACCAGACAGGTTATCCCTGACCTA.................................................................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TCTGATTCTGTGCCTTTCTCTGTCGG................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................TGATTCTGTGCCTTTCTCTGTCGG................................................................ | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 |
| ...............................................................................................................................................................................TTCTCTGTCGGTCCTCTGCCTGCAGGT................................................ | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGATTCTGTGCCTTTCTCTGTCGGT............................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................TTGTTCCATCTGATTCTGTGCCTTTCa....................................................................... | 27 | a | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGTCTTGTTCCATCTGATTCTGTGCCT........................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTGTCGGTCCTCTGCCTGCAGGTGTCt............................................ | 28 | t | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TCTGCCTGCAGGTGTCCTGGTAAAGGT.................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTGTCGGTCCTCTGCCTGCAGGTGT.............................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CAGGGCCAGACTTGAAGTt.......................................................................................................................................................................... | 19 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................TCTGTCGGTCCTCTGCCTGCAGGTG............................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TTCTGTGCCTTTCTCTGTCGGTCCTC........................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TCTTGTTCCATCTGATTCTGTGCCTT.......................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGTGCCTTTCTCTGTCGGTCCTCTGC........................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................CCAGTACAGGGCCAGACTTGAAG............................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................TGGTAAAGGTGGTGGAGGTATACTTC.................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................TCTGTGCCTTTCTCTGTCGGTCCTCTG......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TTCTGTGCCTTTCTCTGTCGGTCCTCTG......................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............TACCAGACAGGTTATCCCTGACCTAT................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGGTAAAGGTGGTGGAGGTATACTTCT................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TTCTCTGTCGGTCCTCTGCCTGCAGGTGTC............................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGATTCTGTGCCTTTCTCTGTCGGTC.............................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGTGTCCTGGTAAAGGTGGTGGA............................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTGTCGGTCCTCTGCCTGCAGGTGTC............................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TAGTCTTGTTCCATCTGATTCTGTGCC............................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTCGGTCCTCTGCCTGCAGGTGTC............................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TCTCTGTCGGTCCTCTGCCTGCAGGT................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGTCGGTCCTCTGCCTGCAGGTGTCCTt.......................................... | 28 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TCTGATTCTGTGCCTTTCT....................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TAAAGGTGGTGGAGGTATACTTCTGT............... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGACATATGCCAGATACCAGACAGGTTATCCCTGACCTATAGGCTTATTATAGCCCAGTACAGGGCCAGACTTGAAGTCTGTCTCTACTCCATGACTATGAAAAGATTCACCTACATTTCTTCTAGTGACTAACTCCTTCAAGTCTTAGTCTTGTTCCATCTGATTCTGTGCCTTTCTCTGTCGGTCCTCTGCCTGCAGGTGTCCTGGTAAAGGTGGTGGAGGTATACTTCTGTGAGCGCTGTGAACAG ..................................................................................................................................................................................(((((.(((.....))).)))))................................................. ...................................................................................................................................................148........................................................207......................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................AGTCTGTCTCTACTggat................................................................................................................................................................ | 18 | ggat | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |