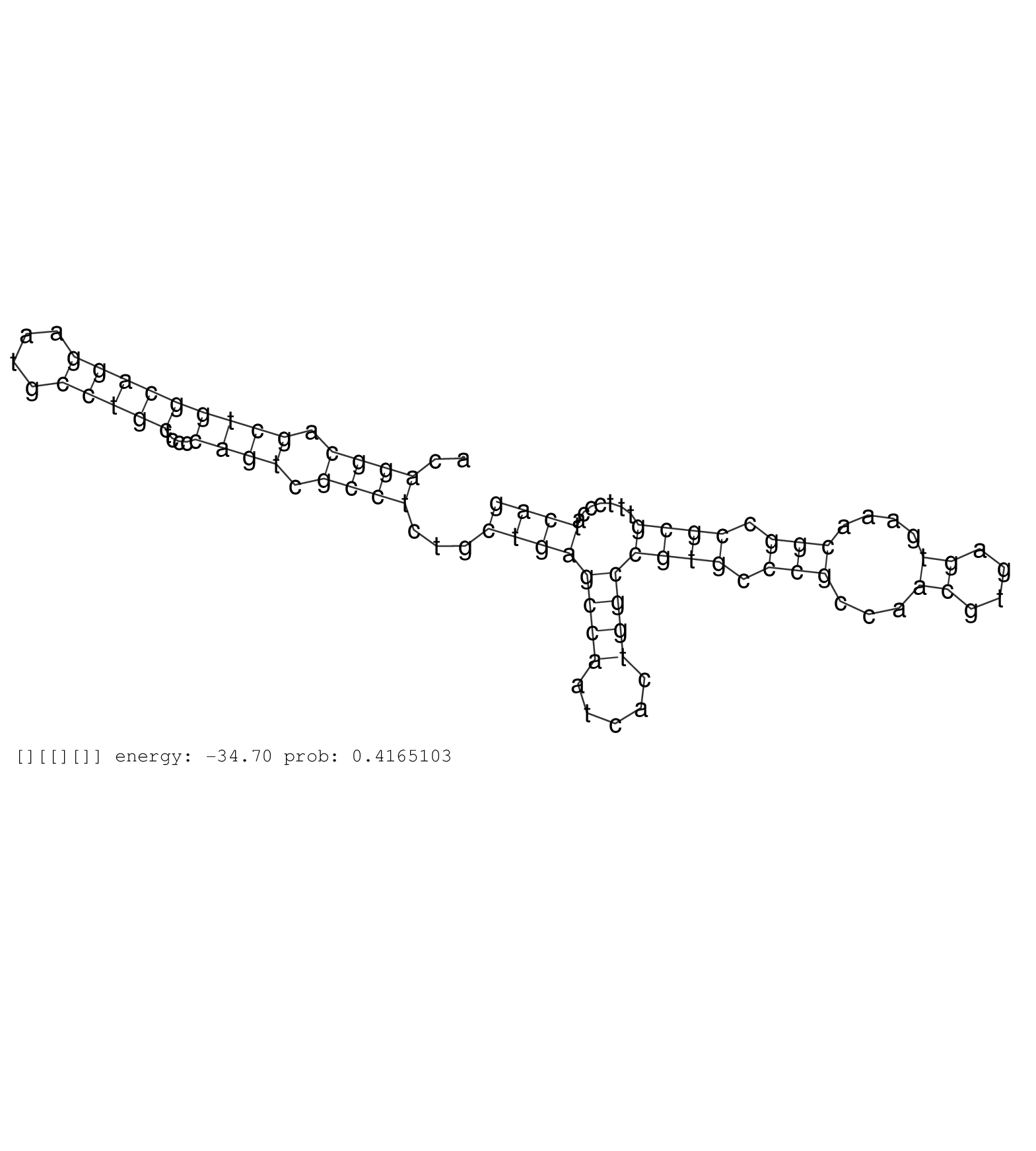

| Gene: Fn1 | ID: uc007bju.1_intron_7_0_chr1_71645038_r.3p | SPECIES: mm9 |
 |
 |
|
(3) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| TGCTCATTCAAGTATCAGCTGTGGTTGTGAAATGACTAACTAGCCAAGGGGCTTTGGGGTTCGCAGAGTAGATAAACAAGGAGTGAAGCTGAAAGACTTCACAGGCAGCTGGCAGGAATGCCTGCTCCCCAGTCGCCTCTGCTGAGCCAATCACTGGCCGTGCCCGCCAACGTGAGTGAAACGGCCGCGTTTCCCATCAGGTCTGGAGCCAGGAACCGAGTACACCATCTACGTCATTGCCCTGAAGAAC ......................................................................................................((((.(((((((((....)))))....)))).))))...((((((((.....))))((((.(((...((....))....))).)))).......)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................AGTAGATAAACAAGGAGTGAAGCTGA.............................................................................................................................................................. | 26 | 1 | 12.00 | 12.00 | - | 6.00 | 2.00 | 3.00 | 1.00 | - | - | - | - | - | - |
| ..................................................................AGTAGATAAACAAGGAGTGAAGCTGAAAG........................................................................................................................................................... | 29 | 1 | 7.00 | 7.00 | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TACGTCATTGCCCTGAAG... | 18 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGTAGATAAACAAGGAGTGAAGCTGAA............................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | - | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - |
| ...................................................................GTAGATAAACAAGGAGTGAAGCTGA.............................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................................................TCTGGAGCCAGGAACCGAGTACACCATCT.................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................................................ATAAACAAGGAGTGAAGCTGAAAG........................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................................................TCTGGAGCCAGGAACCGAGTACAC......................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................AGTAGATAAACAAGGAGTGAAGCTGAAA............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................AGTAGATAAACAAGGAGTGAAGCTGAt............................................................................................................................................................. | 27 | t | 1.00 | 12.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGGAGCCAGGAACCGAGTACACCATa..................... | 26 | a | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................AGTAGATAAACAAGGAGTGAAGCTGt.............................................................................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| TGCTCATTCAAGTATCAGCTGTGGTTGTGAAATGACTAACTAGCCAAGGGGCTTTGGGGTTCGCAGAGTAGATAAACAAGGAGTGAAGCTGAAAGACTTCACAGGCAGCTGGCAGGAATGCCTGCTCCCCAGTCGCCTCTGCTGAGCCAATCACTGGCCGTGCCCGCCAACGTGAGTGAAACGGCCGCGTTTCCCATCAGGTCTGGAGCCAGGAACCGAGTACACCATCTACGTCATTGCCCTGAAGAAC ......................................................................................................((((.(((((((((....)))))....)))).))))...((((((((.....))))((((.(((...((....))....))).)))).......)))).................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................................CTGGCCGTGCCCGCCct.................................................................................. | 17 | ct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |