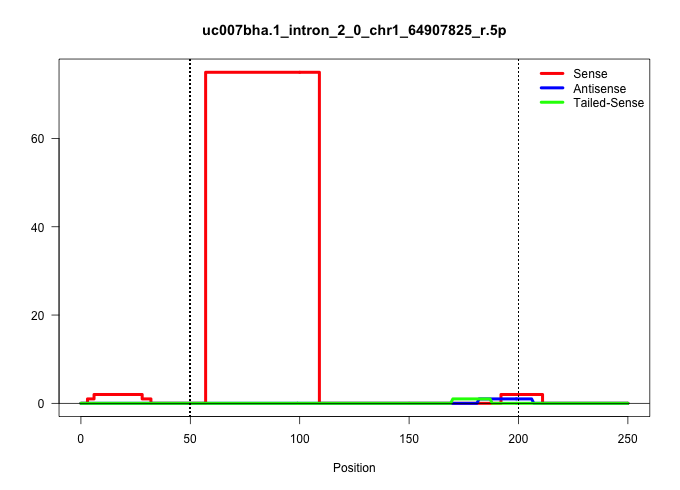
| Gene: 9430067K14Rik | ID: uc007bha.1_intron_2_0_chr1_64907825_r.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(1) PIWI.ip |
(8) TESTES |
| GCCTATTTGTTCAGCTGCCGGGCAGCAGTGGCGGAGGATCTGCGCCGCAGGTAAGTCATCAAATGCCAGCTCAGGAAGCTGGTCAGGTGCCCCCTGAGCCCCTTTCCAAGGTAGCCCTTCTCCAGTCTTTATGTCTCCCGCCACCACCAGACCACCTCTGTCCGTGGCTCATTTCAATTTCTCCTTTGTCCCCAGTCAGTTGAACCAAGCAAAAGTGAGCACCACTGCTCAGCCTTCCCTGCCAGTGTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................ATCAAATGCCAGCTCAGGAAGCTGGTCAGGTGCCCCCTGAGCCCCTTTCCAA............................................................................................................................................. | 52 | 1 | 75.00 | 75.00 | 75.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................CAGTCAGTTGAACCAAGCA....................................... | 19 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - |
| ..........................................................................................................................................................................ATTTCAATTTCTCCTcaa.............................................................. | 18 | caa | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - |
| ...TATTTGTTCAGCTGCCGGGCAGCAG.............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ......TTGTTCAGCTGCCGGGCAGCAGTGGC.......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 |
| GCCTATTTGTTCAGCTGCCGGGCAGCAGTGGCGGAGGATCTGCGCCGCAGGTAAGTCATCAAATGCCAGCTCAGGAAGCTGGTCAGGTGCCCCCTGAGCCCCTTTCCAAGGTAGCCCTTCTCCAGTCTTTATGTCTCCCGCCACCACCAGACCACCTCTGTCCGTGGCTCATTTCAATTTCTCCTTTGTCCCCAGTCAGTTGAACCAAGCAAAAGTGAGCACCACTGCTCAGCCTTCCCTGCCAGTGTGT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................CGCCACCACCAGACCACCcgcg.............................................................................................. | 22 | cgcg | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCTTTGTCCCCAGTCAGTTGAACCA........................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................CGCCACCACCAGACCcgcg................................................................................................. | 19 | cgcg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - |
| ...........................................................ATGCCAGCTCAGGAAGctc............................................................................................................................................................................ | 19 | ctc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................................................CCCCAGTCAGTTGAtt............................................... | 16 | tt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - |