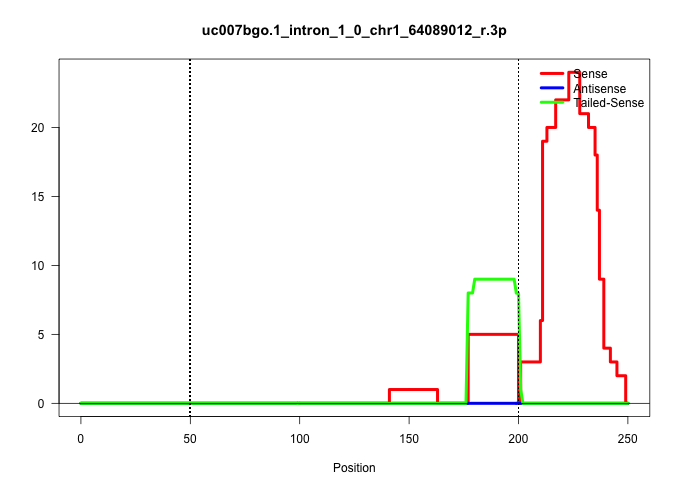
| Gene: Klf7 | ID: uc007bgo.1_intron_1_0_chr1_64089012_r.3p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| CAGCACCAGGGGGCTTTATTTATTATTATTATTATTATTATCATTATTATTATTATTATTTTAAAAGTAATGTTCTATATTAGAGAATGTAGTCTACCAAGAAGGATCTGAGAGGCCATCTGCTGAGACTAGCCACCAGGCAGCAGAACGCAGCGGGCATGAGATCCTTTCTGACCATTGTCCTTCTGTGTCTTCTGCAGGTGAGAAGCCTTACAAGTGCTCATGGGAAGGATGCGAGTGGCGTTTTGCA ...........................................................................................................................................((((.((.((((((.((((((((((.....)))...))).))))..)))))))).)))).................................................... ........................................................................................................................................137............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTGCAG.................................................. | 23 | 1 | 33.00 | 33.00 | 22.00 | - | 4.00 | 1.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTGCAGt................................................. | 24 | t | 12.00 | 33.00 | - | 3.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTGCAGta................................................ | 25 | ta | 6.00 | 33.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TACAAGTGCTCATGGGAAGGATGCGA............. | 26 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TACAAGTGCTCATGGGAAGGATGCGAGT........... | 28 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAAGCCTTACAAGTGCTCATGGGA...................... | 27 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTGCAGa................................................. | 24 | a | 3.00 | 33.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TACAAGTGCTCATGGGAAGGATGCG.............. | 25 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGGGAAGGATGCGAGTGGCGTTTTGC. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................................................................TACAAGTGCTCATGGGAAGGATGC............... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTACAAGTGCTCATGGGAAGGA.................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCCTTCTGTGTCTTCTGCAGt................................................. | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTGCAGtt................................................ | 25 | tt | 1.00 | 33.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGCTCATGGGAAGGATGCGAGTGGC........ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGCAGAACGCAGCGGGCATGAG....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTACAAGTGCTCATGGGAAGGATGCG.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTG..................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTGCt................................................... | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................TTGTCCTTCTGTGTCTTCTGCAtt................................................. | 24 | tt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................CAAGTGCTCATGGGAAGGATGCGA............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................................................................................................................................................................................TGCTCATGGGAAGGATGCGAGTGGCGTT..... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTACAAGTGCTCATGGGAAGGATGCGAGT........... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CTTCTGTGTCTTCTGCAG.................................................. | 18 | 5 | 0.40 | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - | - | - | - | - |
| CAGCACCAGGGGGCTTTATTTATTATTATTATTATTATTATCATTATTATTATTATTATTTTAAAAGTAATGTTCTATATTAGAGAATGTAGTCTACCAAGAAGGATCTGAGAGGCCATCTGCTGAGACTAGCCACCAGGCAGCAGAACGCAGCGGGCATGAGATCCTTTCTGACCATTGTCCTTCTGTGTCTTCTGCAGGTGAGAAGCCTTACAAGTGCTCATGGGAAGGATGCGAGTGGCGTTTTGCA ...........................................................................................................................................((((.((.((((((.((((((((((.....)))...))).))))..)))))))).)))).................................................... ........................................................................................................................................137............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............TTATTATTATTATTATTATTATCATTcagc............................................................................................................................................................................................................. | 30 | cagc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |