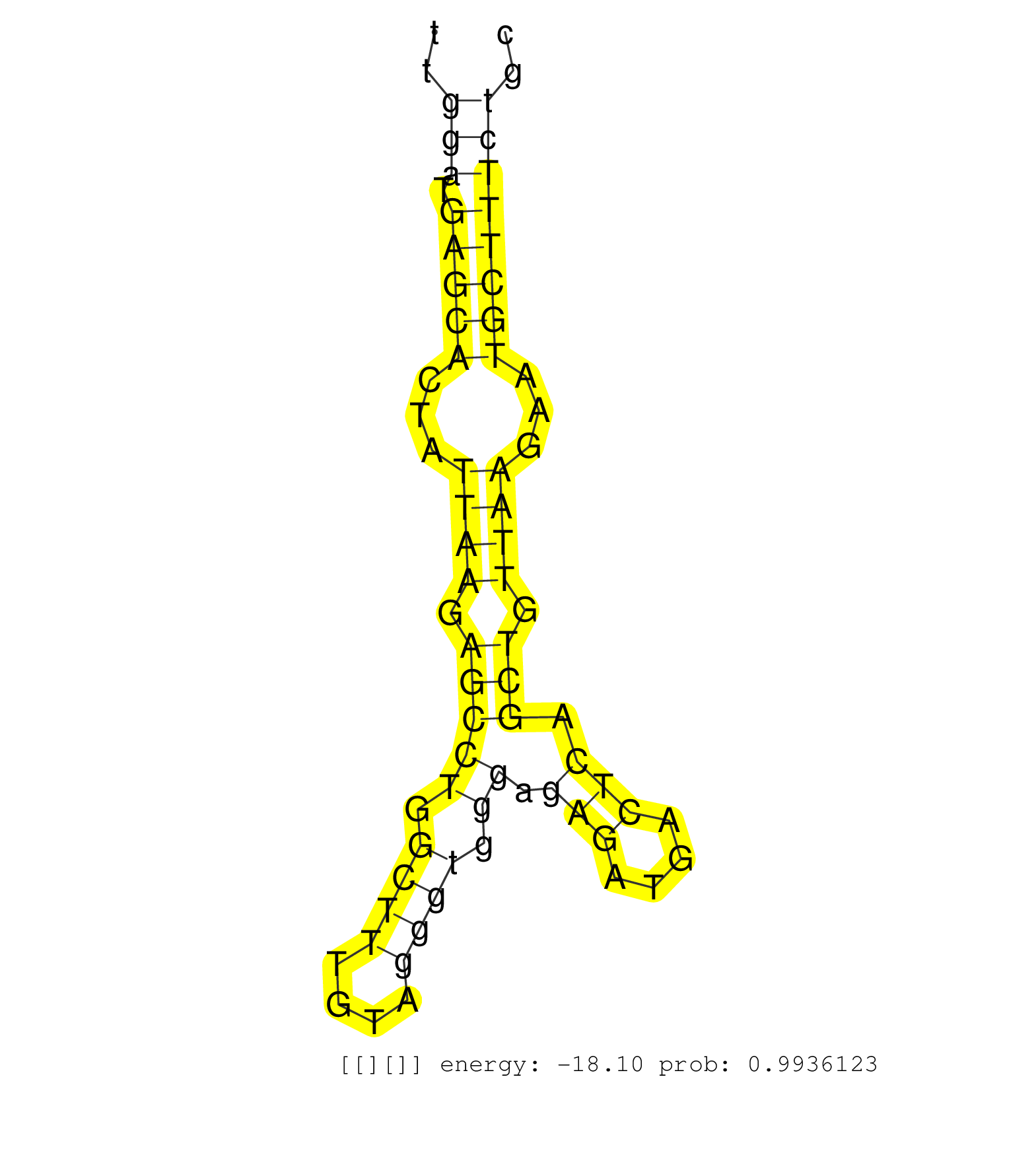
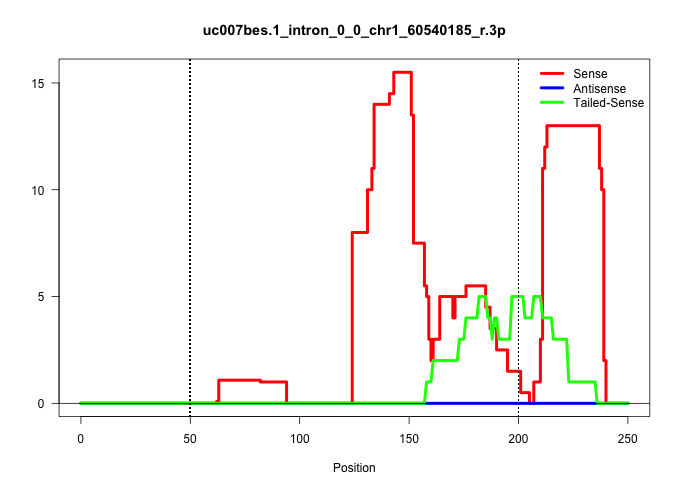
| Gene: Raph1 | ID: uc007bes.1_intron_0_0_chr1_60540185_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(2) OTHER.mut |
(1) OVARY |
(6) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| TGGTCATATAGCCAGAGTAAATTTGTTTTTCCAGTTTCATTATTTTGTCATCTATCCCAATTCTTAATAAAAAAAAAAAAAGTACATAGATTCCATTTAAACAAAGGATTTCAAGTGTTTTGGATGAGCACTATTAAGAGCCTGGCTTTGTAGGGTGGGAGAGATGACTCAGCTGTTAAGAATGCTTTCTGCTCTTCCAGAGGACCAGAGTTCAATTCCCAGCACCCTCGTCGGGCAACTCAAAACCACC .........................................................................................................................(((.(((((...((((.(((((.((((....)))).)).(((....))).))).))))...))))))))............................................................ .......................................................................................................................120.....................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................TGAGCACTATTAAGAGCCTGGCTTTGTA.................................................................................................. | 28 | 1 | 6.00 | 6.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTAAGAGCCTGGCTTTGTAGGGTGGGAGAGATG.................................................................................... | 33 | 1 | 6.00 | 6.00 | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCAATTCCCAGCACCCTCGTCGGGCAAC........... | 28 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCAATTCCCAGCACCCTCGTCGGGCAACT.......... | 29 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGAGGACCAGAGTTCAATTCCCAaa........................... | 26 | aa | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TATTAAGAGCCTGGCTTTGTAGGGTG............................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................TAAGAGCCTGGCTTTGTAGGGTGGG........................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................TGAGCACTATTAAGAGCCTGGCTTTGT................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AGATGACTCAGCTGTTAAGAATGCTTa.............................................................. | 27 | a | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCAATTCCCAGCACCCTCGTCGGGCAA............ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAGTTCAATTCCCAGCACCCTCGTCGGGCA............. | 30 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TAAGAGCCTGGCTTTGTAGGGTGGGA.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAAGAATGCTTTCTGCTCTTCCAGAGt............................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................AGAGATGACTCAGCTGTTAAGAATGC................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AATTCCCAGCACCCTCGTCGGGCAAC........... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TGCTTTCTGCTCTTCCAGAGGACCAGAtt....................................... | 29 | tt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGACTCAGCTGTTAAGAATGCTTTCT............................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TGACTCAGCTGTTAAGAATGCTTTCTGCTCT....................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................................GAGAGATGACTCAGCTGTTAAGAAccgt................................................................ | 28 | ccgt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTCAATTCCCAGCACCCTCGTCGGGCAAC........... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CAATTCCCAGCACCCTCGTCGGGCAAC........... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GAGTTCAATTCCCAGCACCCTCGTCGGaa.............. | 29 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTAAGAGCCTGGCTTTGTAGGGTGGG........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GGCTTTGTAGGGTGGGAGAGATGACTC................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TTAATAAAAAAAAAAAAAGTACATAGATTCC............................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AGATGACTCAGCTGTTAAGAATGCTT............................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TTCAATTCCCAGCACCCTCGTCGGGCA............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGTTAAGAATGCTTgtca........................................................... | 18 | gtca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................GCTGTTAAGAATGCTTTCTGCTCTTCCAGA................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................TGCTCTTCCAGAGGACCAGAGTTCAta.................................. | 27 | ta | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAAGAATGCTTTCTGCTCTTCCAGAGGAC............................................. | 29 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CTGGCTTTGTAGGGTGG............................................................................................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................CTTAATAAAAAAAAAAAAAG........................................................................................................................................................................ | 20 | 12 | 0.08 | 0.08 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.08 |
| TGGTCATATAGCCAGAGTAAATTTGTTTTTCCAGTTTCATTATTTTGTCATCTATCCCAATTCTTAATAAAAAAAAAAAAAGTACATAGATTCCATTTAAACAAAGGATTTCAAGTGTTTTGGATGAGCACTATTAAGAGCCTGGCTTTGTAGGGTGGGAGAGATGACTCAGCTGTTAAGAATGCTTTCTGCTCTTCCAGAGGACCAGAGTTCAATTCCCAGCACCCTCGTCGGGCAACTCAAAACCACC .........................................................................................................................(((.(((((...((((.(((((.((((....)))).)).(((....))).))).))))...))))))))............................................................ .......................................................................................................................120.....................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT1() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................CCAGAGGACCAGAGTTCAATtt.................................. | 22 | tt | 4.00 | 0.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................CTGCTCTTCCAGAGGACCAgta........................................... | 22 | gta | 2.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TTCCAGAGGACCAGAGTTCAATcag.................................. | 25 | cag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...........................................................................................................................................................................................TGCTCTTCCAGAGGACCAGAGTTCAaa.................................... | 27 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TCTTCCAGAGGACCAGAGTTCAAaa................................... | 25 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |