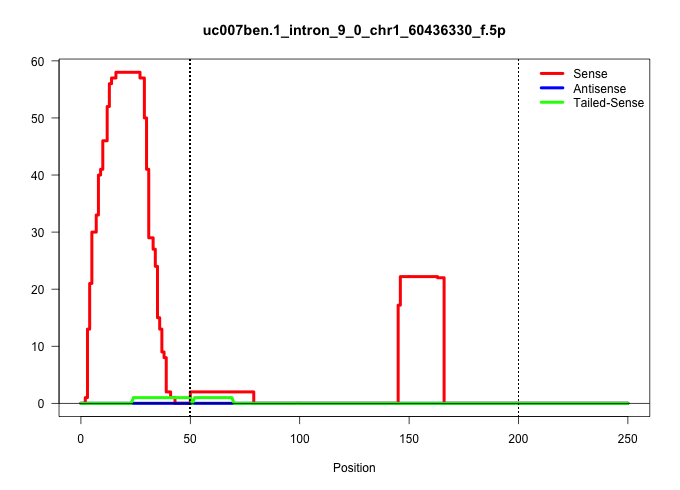
| Gene: Cyp20a1 | ID: uc007ben.1_intron_9_0_chr1_60436330_f.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| GGCTTCAAGACATTGAAGGAAAAGTCGGCCCATTTGTTATTCCTAAGGAGGTAGGAAGTGTTTGGTATATTTAACTTAGTCAGTTTATCCTCAACTACTTTGGTATGGGCCTTTATTTTATTTCTTTTCATTGAAAATCACTTTTTTCATATGATGTATTCTGATCACAGTTTCTCCTCCCTCATCTGTTCCCATATCCTCCCTTCCTTCCCACTCCATGGTCTTCCTTCTTTCTTTAGAAAACAAACTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................TTCATATGATGTATTCTGATC.................................................................................... | 21 | 1 | 17.00 | 17.00 | - | - | - | - | 6.00 | 5.00 | 2.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .....CAAGACATTGAAGGAAAAGTCGGCCC........................................................................................................................................................................................................................... | 26 | 1 | 7.00 | 7.00 | 1.00 | - | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ........GACATTGAAGGAAAAGTCGGCCCATTT....................................................................................................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...TTCAAGACATTGAAGGAAAAGTCGGC............................................................................................................................................................................................................................. | 26 | 1 | 6.00 | 6.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................TCATATGATGTATTCTGATC.................................................................................... | 20 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 2.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - |
| ....TCAAGACATTGAAGGAAAAGTCGGCC............................................................................................................................................................................................................................ | 26 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............TGAAGGAAAAGTCGGCCCATTTGTTA................................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ...TTCAAGACATTGAAGGAAAAGTCGGCC............................................................................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTGAAGGAAAAGTCGGCCCATTTGTTA................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...TTCAAGACATTGAAGGAAAAGTCGGCCC........................................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TTGAAGGAAAAGTCGGCCCATTTGT..................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - |
| ....TCAAGACATTGAAGGAAAAGTCGGCCC........................................................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CATTGAAGGAAAAGTCGGCCCATTTG...................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGAAGTGTTTGGTATATTTAACTTAG........................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......AGACATTGAAGGAAAAGTCGGCCCAT......................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GAAGGAAAAGTCGGCCCATTTGT..................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........CATTGAAGGAAAAGTCGGCCCATTT....................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............TGAAGGAAAAGTCGGCCCATTTGTTATT................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CAAGACATTGAAGGAAAAGTCGGCCCATT........................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........GACATTGAAGGAAAAGTCGGCCCATT........................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CATTGAAGGAAAAGTCGGCCCATT........................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............TTGAAGGAAAAGTCGGCCCATTTGTT.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........CATTGAAGGAAAAGTCGGCCCATTTGT..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....CAAGACATTGAAGGAAAAGTCGGCCCATTT....................................................................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGAAGTGTTTGGTgagg.................................................................................................................................................................................... | 18 | gagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................AGGAAAAGTCGGCCCATTTGTTATTCC............................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........ACATTGAAGGAAAAGTCGGCCCATTT....................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................TCGGCCCATTTGTTATTCCTAAGGAGa....................................................................................................................................................................................................... | 27 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......AGACATTGAAGGAAAAGTCG............................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..CTTCAAGACATTGAAGGAAAAGTCGGCC............................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....TCAAGACATTGAAGGAAAAGTCGGC............................................................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TTCATATGATGTATTCTG....................................................................................... | 18 | 5 | 0.20 | 0.20 | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - |
| GGCTTCAAGACATTGAAGGAAAAGTCGGCCCATTTGTTATTCCTAAGGAGGTAGGAAGTGTTTGGTATATTTAACTTAGTCAGTTTATCCTCAACTACTTTGGTATGGGCCTTTATTTTATTTCTTTTCATTGAAAATCACTTTTTTCATATGATGTATTCTGATCACAGTTTCTCCTCCCTCATCTGTTCCCATATCCTCCCTTCCTTCCCACTCCATGGTCTTCCTTCTTTCTTTAGAAAACAAACTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|