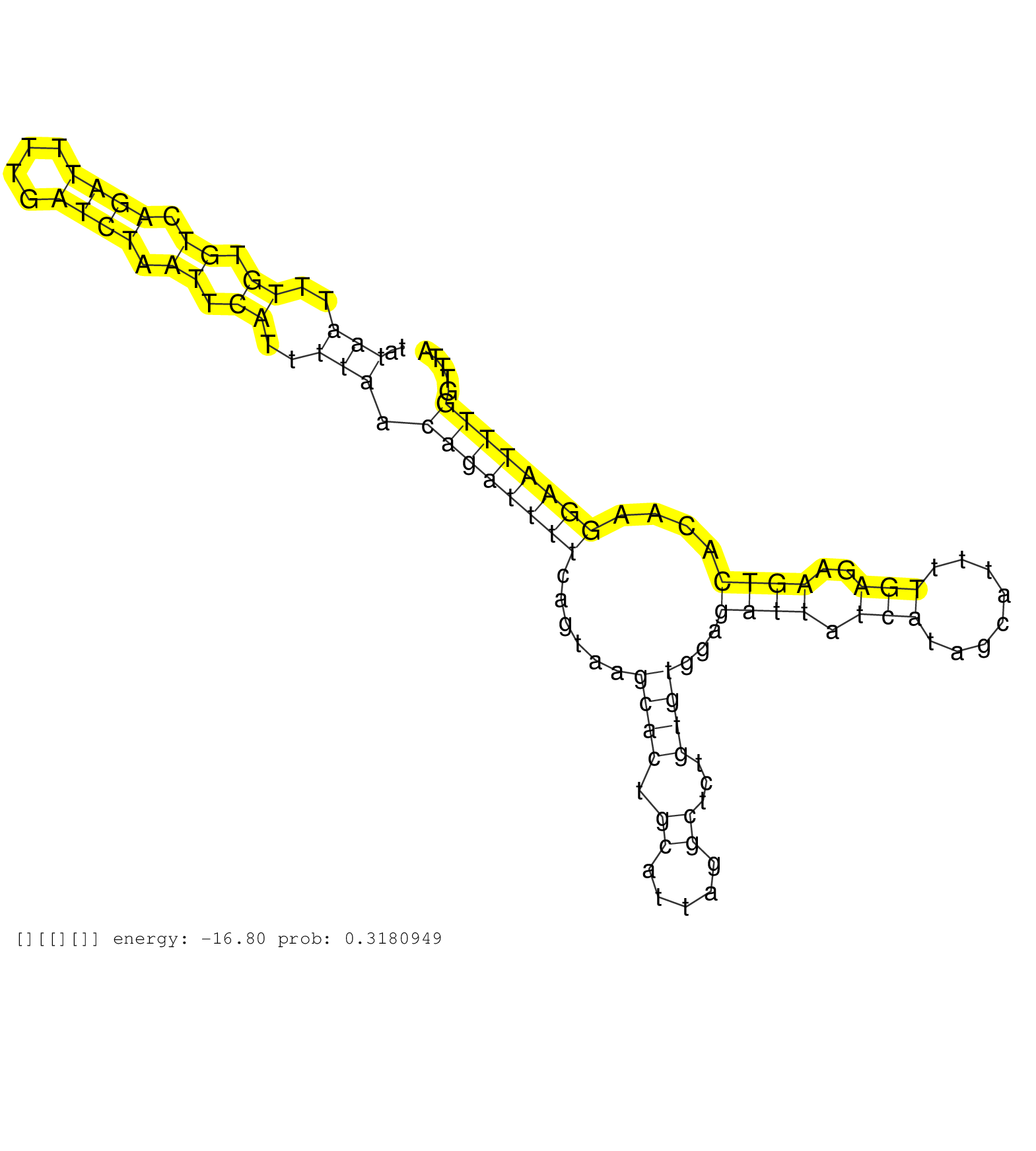

| Gene: Nabp1 | ID: uc007axm.1_intron_0_0_chr1_51525931_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(12) TESTES |
| CTCCCTAACTCTGTACCTGTCATTTCTCCTTACCTCATATTGTAAAATAAGTAAGTTTCTGTTCAACTTGTATAGCATTTGAATATAATTTGTGTCAGATTTTGATCTAATTCATTTTAACAGATTTTCAGTAAGCACTGCATTAGGCTCTGTGTGGAGATTATCATAGCATTTTGAGAAGTCACAAGGAATTTGGTTTATAGGTACAGGGCTTTGACATTTGTCATTGTTCATATATTAATAATTATAG .....................................................................................(((..((.((.((((....)))).)).))..))).((((((((......((((.((.....))...))))...((((.(((........)))..))))....))))))))....................................................... ...................................................................................84..................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................TGAGAAGTCACAAGGAATTTGGTTTA.................................................. | 26 | 1 | 9.00 | 9.00 | 2.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TTTGAGAAGTCACAAGGAATTTGGTTT................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAAGTCACAAGGAATTTGGTTTAT................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAAGTCACAAGGAATTTGGTTT................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................TTTGTGTCAGATTTTGATCTAATTCAT....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................GTGGAGATTATCATAGCATTTTGAGAA...................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....CTAACTCTGTACCTGTCATTTCTCCTT........................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................CATTTGAATATAATTTGTGTC.......................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................TAATTTGTGTCAGATTTTGATCTAATT.......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGCATTAGGCTCTGTGTGGAGATTATCA.................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGCATTAGGCTCTGTGTGGAGATTA....................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGCATTAGGCTCTGTGTGGAGATT........................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................TTAGGCTCTGTGTGGAGATTATCATA.................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..CCCTAACTCTGTACCTGTCATTTCTCC............................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................ACAGATTTTCAGTAAGCACTGCATTAGGCTC.................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................TATAATTTGTGTCAGATTTTGATCTAA............................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAAGTCACAAGGAATTTGGTTTATAt............................................... | 29 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGAGAAGTCACAAGGAATTTGGTTTATAGt.............................................. | 30 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................TCACAAGGAATTTGGTTTATAGGTACAGG........................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................AAGCACTGCATTAGGCTCTGTGTGGAG........................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........CTCTGTACCTGTCATTTCTCC............................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................................ACTGCATTAGGCTCTGTGTGGAGATTATC..................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TCTGTTCAACTTGTATAGCATTTGAATA..................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................AGTAAGCACTGCATTAGGCTCTGTGT............................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................GTACAGGGCTTTGAC................................ | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| CTCCCTAACTCTGTACCTGTCATTTCTCCTTACCTCATATTGTAAAATAAGTAAGTTTCTGTTCAACTTGTATAGCATTTGAATATAATTTGTGTCAGATTTTGATCTAATTCATTTTAACAGATTTTCAGTAAGCACTGCATTAGGCTCTGTGTGGAGATTATCATAGCATTTTGAGAAGTCACAAGGAATTTGGTTTATAGGTACAGGGCTTTGACATTTGTCATTGTTCATATATTAATAATTATAG .....................................................................................(((..((.((.((((....)))).)).))..))).((((((((......((((.((.....))...))))...((((.(((........)))..))))....))))))))....................................................... ...................................................................................84..................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................CTCCTTACCTCATATtt.................................................................................................................................................................................................................. | 17 | tt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |