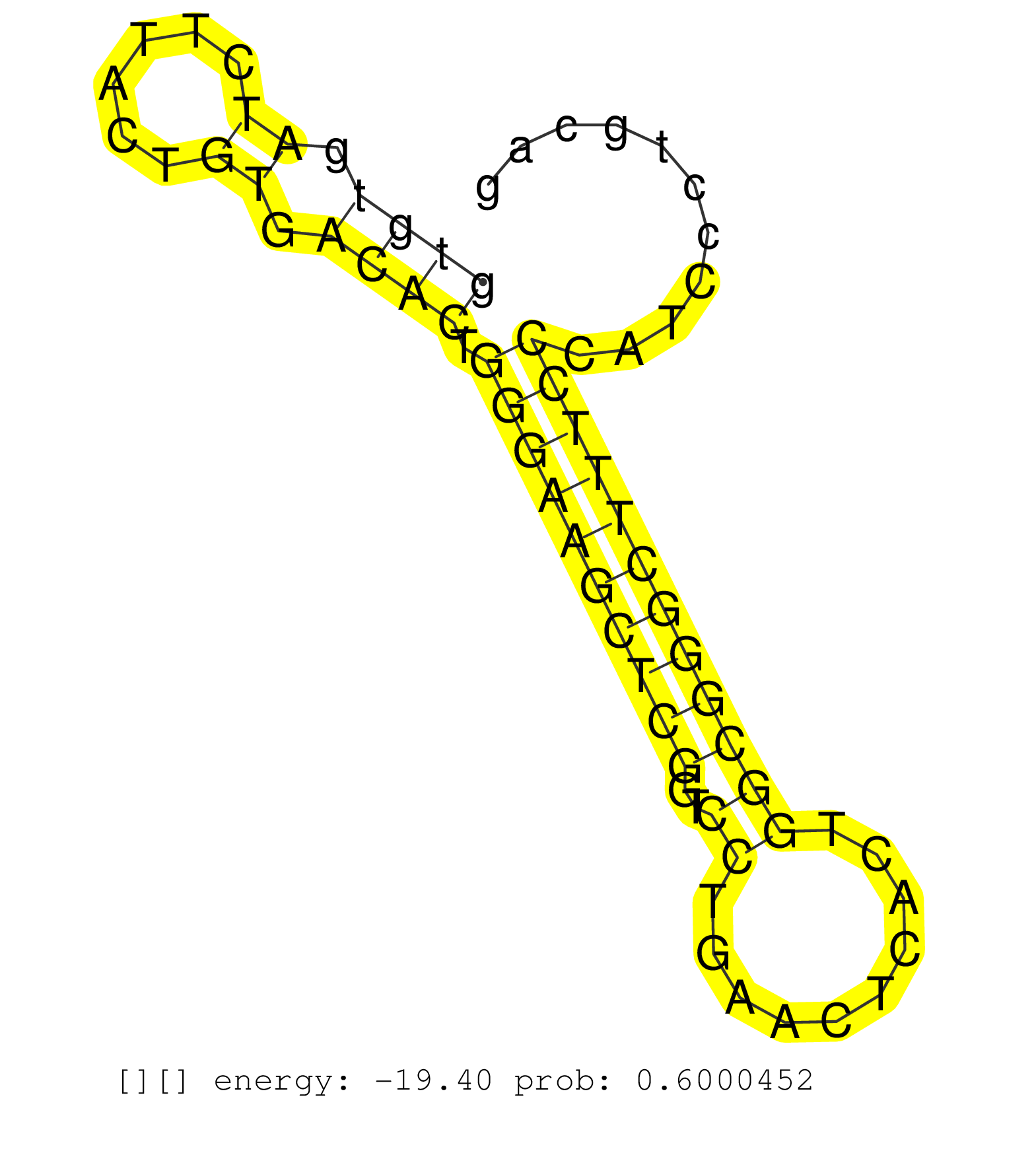
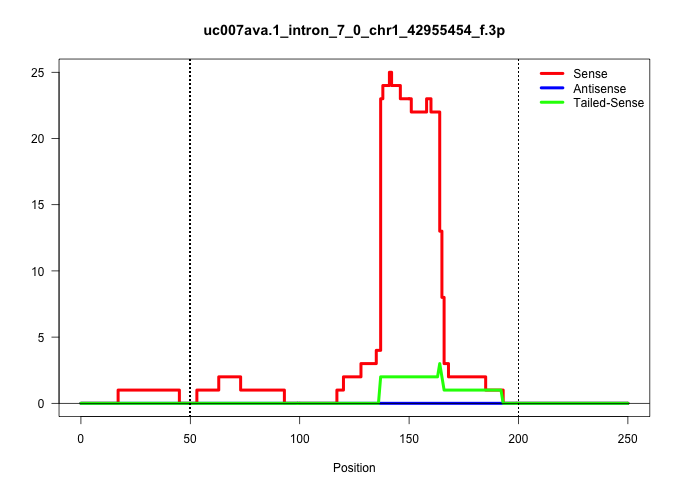
| Gene: Mrps9 | ID: uc007ava.1_intron_7_0_chr1_42955454_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(17) TESTES |
| CATGGGTGTTTCTGAGATGGAATTGCAGAGCATAATTCCCCCACAGTCTGTCGGCCTCATGACTCAGGCGTAACTTTTGACTATAGTAGTCGTGGAAGTTTCCTCTTTGCAGGCTTATGCAGTCAGCGGGAAGTGTGATCTTACTGTGACACTGGGAAGCTCGGTCCTGAACTCACTGGCGGGCTTTCCCATCCCTGCAGGCCGGAGGAAGAGCGCCACGGCACAGGCGGTCGTTTATGAGCACGGAAGC ....................................................................................................................................((((.((......)).)))).((((((((((..((..........))))))))))))............................................................. ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................ATCTTACTGTGACACTGGGAAGCTCGG...................................................................................... | 27 | 1 | 8.00 | 8.00 | 2.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATCTTACTGTGACACTGGGAAGCTCGGT..................................................................................... | 28 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATCTTACTGTGACACTGGGAAGCTCGGTC.................................................................................... | 29 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................ACAGGCGGTCGTTTATGAGCACGGAAG. | 27 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................GCTCGGTCCTGAACTCACTGGCGGGCT................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................AGTCAGCGGGAAGTGTGATCTTACTG........................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................TGGAATTGCAGAGCATA........................................................................................................................................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................TCCTGAACTCACTGGCGGGCTTTCCCATt......................................................... | 29 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGGAATTGCAGAGCATAATTCCCCCACA............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................TACTGTGACACTGGGAAGCTCGGTC.................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................TCAGGCGTAACTTTTGACTATAGTAGTCGT............................................................................................................................................................. | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................GGAAGTGTGATCTTACTGTGACA................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................TGATCTTACTGTGACACTGGGAAGC.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CCGGAGGAAGAGCGCCACGGCACAGGC...................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................CTGAACTCACTGGCGGGCTTTCCCATC......................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TCTTACTGTGACACTGGGAAGCTCGG...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................ATCTTACTGTGACACTGGGAAGCTCGGTt.................................................................................... | 29 | t | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GCCTCATGACTCAGGCGTAA................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CAGGCGGTCGTTTATGAGCACGGAAGC | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATCTTACTGTGACACTGGGAAGCTCGGTCCT.................................................................................. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .....................................................................................................................TGCAGTCAGCGGGAAGTGTGATCTT............................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................ATCTTACTGTGACACTGGGAAGCTCGGa..................................................................................... | 28 | a | 1.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CATGGGTGTTTCTGAGATGGAATTGCAGAGCATAATTCCCCCACAGTCTGTCGGCCTCATGACTCAGGCGTAACTTTTGACTATAGTAGTCGTGGAAGTTTCCTCTTTGCAGGCTTATGCAGTCAGCGGGAAGTGTGATCTTACTGTGACACTGGGAAGCTCGGTCCTGAACTCACTGGCGGGCTTTCCCATCCCTGCAGGCCGGAGGAAGAGCGCCACGGCACAGGCGGTCGTTTATGAGCACGGAAGC ....................................................................................................................................((((.((......)).)))).((((((((((..((..........))))))))))))............................................................. ....................................................................................................................................133................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|