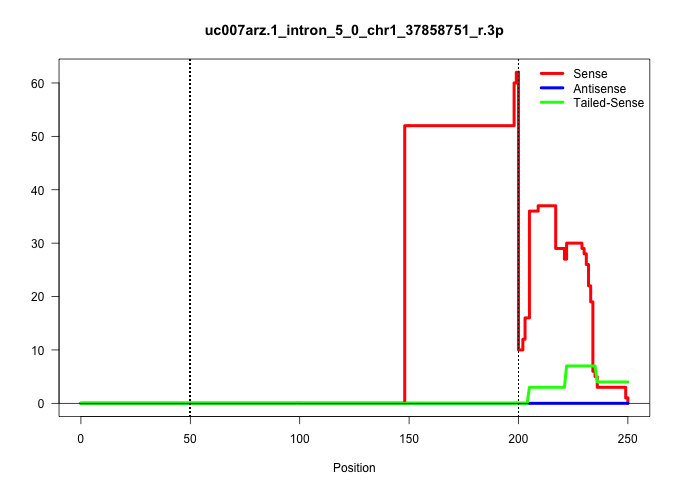
| Gene: Tsga10 | ID: uc007arz.1_intron_5_0_chr1_37858751_r.3p | SPECIES: mm9 |
 |
|
(3) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| AGGAACTGATGGACGCAGGTTGGGGCATTCTTCTTTATGTTCCTTACAGGTTGACCATCTTTCTAGGCTAAGGGGTTAGTAGGATTTCTTCAGGGTTATGTACCAGTGATTTCCCAGTGGCCTAGATAGCAGTGTTCCAAAAGCACTTTATGTTAAATACTTTTAAAGTTTTAAAGTAATACACAAGATTTGCTTTTTAGGAATCTGAGAACCGGCAAATAATGGAACAACTCCGAAAAGCCAATGAAGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................TATGTTAAATACTTTTAAAGTTTTAAAGTAATACACAAGATTTGCTTTTTAG.................................................. | 52 | 1 | 52.00 | 52.00 | 52.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAGAACCGGCAAATAATGGAACAACTCC................ | 29 | 1 | 13.00 | 13.00 | - | 6.00 | 6.00 | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGAATCTGAGAACCGGCA................................. | 19 | 1 | 8.00 | 8.00 | - | - | - | 8.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGAACAACTCCGAAAAGCCAATGAAGAtt | 30 | tt | 4.00 | 1.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAGAACCGGCAAATAATGGAACAACTCCGt.............. | 31 | t | 3.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................TGAGAACCGGCAAATAATGGAACAACTC................. | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAGAACCGGCAAATAATGGAACAACT.................. | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGAACAACTCCGAAAAGCCAATGAAG. | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................................................................................................................................TCTGAGAACCGGCAAATAATGGAACAAC................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAATCTGAGAACCGGCAAATA............................. | 22 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TGAGAACCGGCAAATAATGGAACAACTCCGA.............. | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGGAACAACTCCGAAAAGCCAATGAAGA | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................TCTGAGAACCGGCAAATAATGGAACAACT.................. | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ATCTGAGAACCGGCAAATAATGGAACAACT.................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................................................................................TGAGAACCGGCAAATAATGGAACAACTCCG............... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AACCGGCAAATAATGGAACAACTCCGA.............. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................................................................TCTGAGAACCGGCAAATAATGGAACAA.................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................ATCTGAGAACCGGCAAATAATGGAACA..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| AGGAACTGATGGACGCAGGTTGGGGCATTCTTCTTTATGTTCCTTACAGGTTGACCATCTTTCTAGGCTAAGGGGTTAGTAGGATTTCTTCAGGGTTATGTACCAGTGATTTCCCAGTGGCCTAGATAGCAGTGTTCCAAAAGCACTTTATGTTAAATACTTTTAAAGTTTTAAAGTAATACACAAGATTTGCTTTTTAGGAATCTGAGAACCGGCAAATAATGGAACAACTCCGAAAAGCCAATGAAGA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|