
| Gene: Inpp4a | ID: uc007ark.1_intron_1_0_chr1_37414250_f | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(9) TESTES |
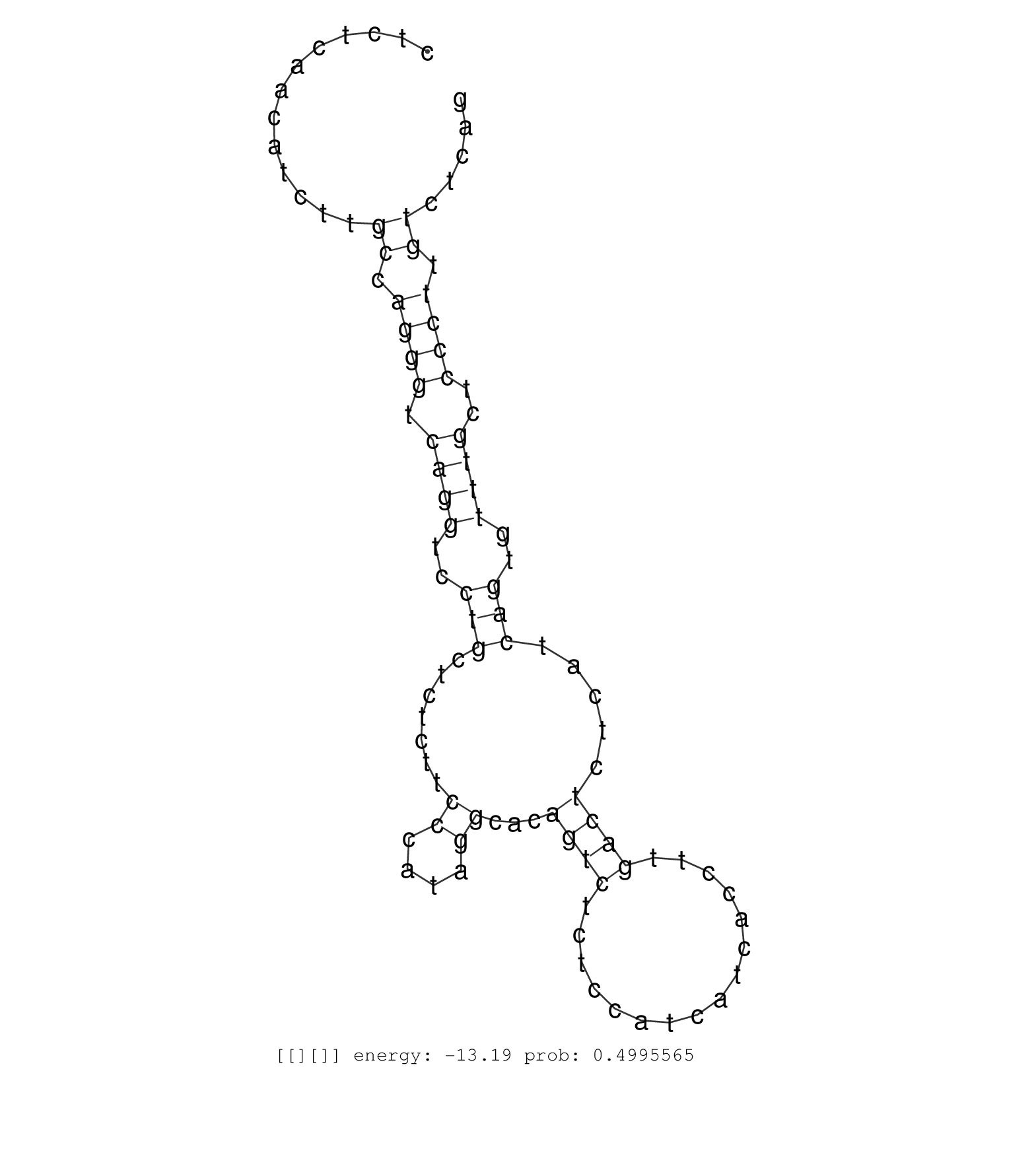
| GATGTCCTTCTGTGACGCAATGCCACGTGGTCCAAAAGCAAGCATGTATGGTAAGTCACTTGTCAGACTGACTTGAGCAGTAGCCTGGCTCTTCACATAGTCTCTGTCCTTCCAGCAGAGGTTATGCTGTGGGACTTGATAGAAGATTGAAGATTCTGTGAGACTTGCCAGTAGAAGAGTGCTAGAGAGCAGTAAGAAACTCATAAACACATTACTCTACAGCTACATTTAGGCAGTCTGTAATGCAGCCTGTTTTGGTAGAAGAGCATCCTCGGGTACACCATGCAGAGGGCAGGGAGGAGGAAGACAGGAGGAGGTGTCAGACATCCTAGCTCAATTCCACCCCGTCACAAGACACTGCTCTCAACATCTTGCCAGGGTCAGGTCCTGCTCTCTTCCCATAGGCACAGTCTCTCCATCATCACCTTGACTCTCATCAGTGTTTGCTCCCTTGTCTCAGGGCTACTGCCATCGTAGGAGTCCAGGGCCTGGAGACGGCTCTGCTTTGAG .....................................................................................................................................................................................................................................................................................................................................................................................((.((((.((((..(((.......((....))...((((................)))).....)))..))))..)))).))....................................................... ........................................................................................................................................................................................................................................................................................................................................................................361................................................................................................460................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................TCTGTCCTTCCAGCAGAGGTTATGCTGTGGGACTTGATAGAAGATTGAAGAT.................................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 72.00 | 72.00 | 72.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CGTAGGAGTCCAGGGCCTGGAGACGGC........... | 27 | 1 | 8.00 | 8.00 | - | 8.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CTGCCATCGTAGGAGTCCAGGGCCTGGAGACG............. | 32 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................................................TTTTGGTAGAAGAGCAgaga.............................................................................................................................................................................................................................................. | 20 | gaga | 3.00 | 0.00 | - | - | 2.00 | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................................................................................................................................................................AGGAGGTGTCAGACAag...................................................................................................................................................................................... | 17 | ag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGCCATCGTAGGAGTCCAGGGC...................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| .......TTCTGTGACGCAATGCCACGTGGTCC............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................GAGGAGGTGTCAGAaaag...................................................................................................................................................................................... | 18 | aaag | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - |
| ....................TGCCACGTGGTCCAAAAGCAAGCATG................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................................................................TTTTGGTAGAAGAGCAgcga.............................................................................................................................................................................................................................................. | 20 | gcga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |
| GATGTCCTTCTGTGACGCAATGCCACGTGGTCCAAAAGCAAGCATGTATGGTAAGTCACTTGTCAGACTGACTTGAGCAGTAGCCTGGCTCTTCACATAGTCTCTGTCCTTCCAGCAGAGGTTATGCTGTGGGACTTGATAGAAGATTGAAGATTCTGTGAGACTTGCCAGTAGAAGAGTGCTAGAGAGCAGTAAGAAACTCATAAACACATTACTCTACAGCTACATTTAGGCAGTCTGTAATGCAGCCTGTTTTGGTAGAAGAGCATCCTCGGGTACACCATGCAGAGGGCAGGGAGGAGGAAGACAGGAGGAGGTGTCAGACATCCTAGCTCAATTCCACCCCGTCACAAGACACTGCTCTCAACATCTTGCCAGGGTCAGGTCCTGCTCTCTTCCCATAGGCACAGTCTCTCCATCATCACCTTGACTCTCATCAGTGTTTGCTCCCTTGTCTCAGGGCTACTGCCATCGTAGGAGTCCAGGGCCTGGAGACGGCTCTGCTTTGAG .....................................................................................................................................................................................................................................................................................................................................................................................((.((((.((((..(((.......((....))...((((................)))).....)))..))))..)))).))....................................................... ........................................................................................................................................................................................................................................................................................................................................................................361................................................................................................460................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................GCAGTCTGTAATGCAGag...................................................................................................................................................................................................................................................................... | 18 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................GTCAGACTGACTTGA.................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 15 | 4 | 0.25 | 0.25 | - | - | - | - | 0.25 | - | - | - | - |