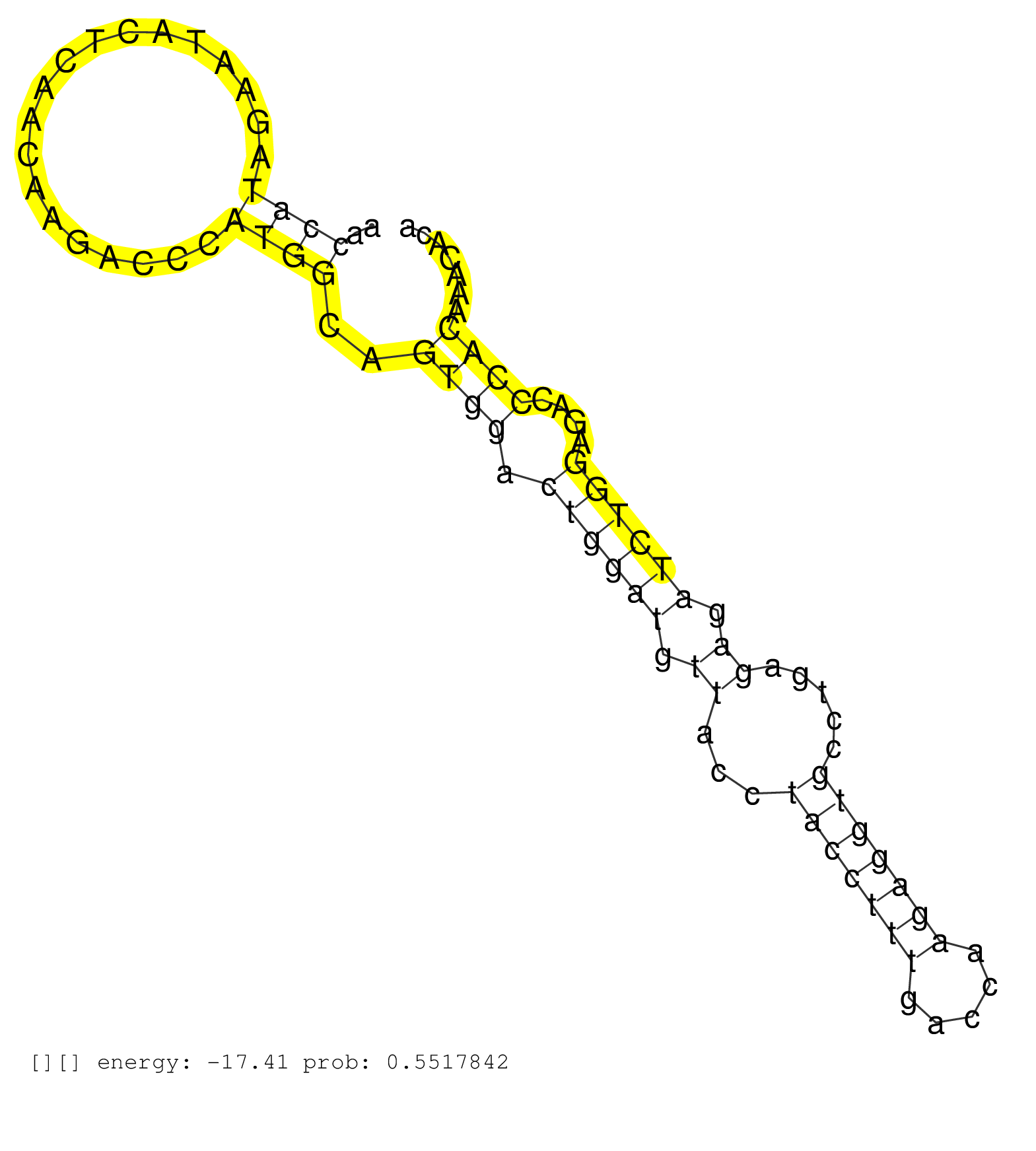
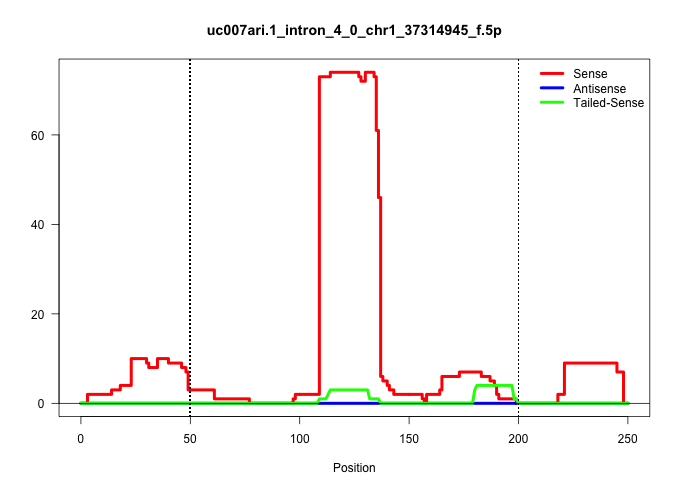
| Gene: Cnga3 | ID: uc007ari.1_intron_4_0_chr1_37314945_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| CTCTGCAGATGTCCTTTATGTTTTAGACATGCTGGTTCGAGCCCGGACAGGTGAGTGTGCCCCACGGACTTGACTGGTCCAGGTCACAGGCCCTCAGTAGGGGGAACCATAGAATACTCAACAAGACCCATGGCAGTGGACTGGATGTTACCTACCTTTGACCAAGAGGTGCCTGAGAGATCTGGAGACCCACAAACACAGAGTGGGTGCTGGTATCTTGATACTGAATCCTTCAACAAGACCTGGTCAG ..........................................................................................................((((...................))))..((((.((((((.((...(((((((.....))))))).....)).))))))....))))......................................................... ........................................................................................................105............................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................TAGAATACTCAACAAGACCCATGGCAGT................................................................................................................. | 28 | 1 | 40.00 | 40.00 | 16.00 | 9.00 | 4.00 | 3.00 | - | - | 3.00 | - | 1.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................TAGAATACTCAACAAGACCCATGGCAG.................................................................................................................. | 27 | 1 | 16.00 | 16.00 | 4.00 | 4.00 | 1.00 | 3.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| .............................................................................................................TAGAATACTCAACAAGACCCATGGCA................................................................................................................... | 26 | 1 | 12.00 | 12.00 | 3.00 | 4.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TACTGAATCCTTCAACAAGACCTGGTC.. | 27 | 1 | 7.00 | 7.00 | - | - | 5.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TAGACATGCTGGTTCGAGCCCGGACA......................................................................................................................................................................................................... | 26 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCTGGAGACCCACAAAgt.................................................... | 18 | gt | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................TTCGAGCCCGGACAGGTGAGTGTGCC............................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGATACTGAATCCTTCAACAAGACCTG..... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GAGGTGCCTGAGAGATCTGGAGAC............................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TAGAATACTCAACAAGACCCATGGCAGg................................................................................................................. | 28 | g | 1.00 | 16.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................AGGGGGAACCATAGAATACTCAACAAGAC........................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGGCAGTGGACTGGATGTTACCTACC.............................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TACTCAACAAGACCCATGGCAGTGGACTG........................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGAGAGATCTGGAGACCCACAAACAC................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGCAGATGTCCTTTATGTTTTAGACATG........................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTGCCCCACGGACTTGACTGG............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...TGCAGATGTCCTTTATGTTTTAGACAT............................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TGACCAAGAGGTGCCTGAGAGATCT................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TAGACATGCTGGTTCGAGCCCGGACAG........................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................AGAGGTGCCTGAGAGATCTGGAGACC............................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGGCAGTGGACTGGATGTTACCTACCT............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TACTCAACAAGACCaaga...................................................................................................................... | 18 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................TAGAATACTCAACAAGACCCATGGCAGTG................................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CTGGAGACCCACAAAgagc.................................................. | 19 | gagc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................TAGAATACTCAACAAGACCCATGGCAGTGGA.............................................................................................................. | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TAGGGGGAACCATAGAATACTCAACAAGACC.......................................................................................................................... | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GAGGTGCCTGAGAGATCTGGAGACC............................................................ | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................GAGGTGCCTGAGAGATCTGGAGACCC........................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TGACCAAGAGGTGCCTGAGAGATCTGGAG............................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TTTATGTTTTAGACATGCTGGTTCGA.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TAGAATACTCAACAAGACCCATGGC.................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TAGACATGCTGGTTCGAGCCCGGAC.......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TGTTTTAGACATGCTGGTTCGAGCCCGG............................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TAGAATACTCAACAAGACCCATGGCAGTGGAC............................................................................................................. | 32 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................ATACTCAACAAGACCaaga...................................................................................................................... | 19 | aaga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| CTCTGCAGATGTCCTTTATGTTTTAGACATGCTGGTTCGAGCCCGGACAGGTGAGTGTGCCCCACGGACTTGACTGGTCCAGGTCACAGGCCCTCAGTAGGGGGAACCATAGAATACTCAACAAGACCCATGGCAGTGGACTGGATGTTACCTACCTTTGACCAAGAGGTGCCTGAGAGATCTGGAGACCCACAAACACAGAGTGGGTGCTGGTATCTTGATACTGAATCCTTCAACAAGACCTGGTCAG ..........................................................................................................((((...................))))..((((.((((((.((...(((((((.....))))))).....)).))))))....))))......................................................... ........................................................................................................105............................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................GAATACTCAACAAGACcat........................................................................................................................... | 19 | cat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |