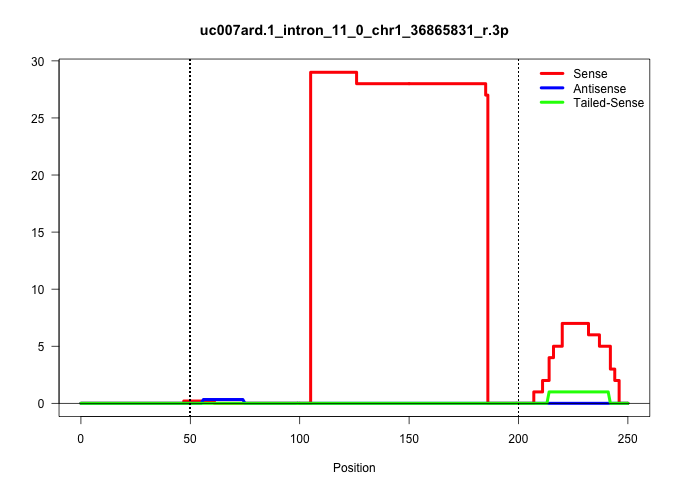
| Gene: Tmem131 | ID: uc007ard.1_intron_11_0_chr1_36865831_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(10) TESTES |
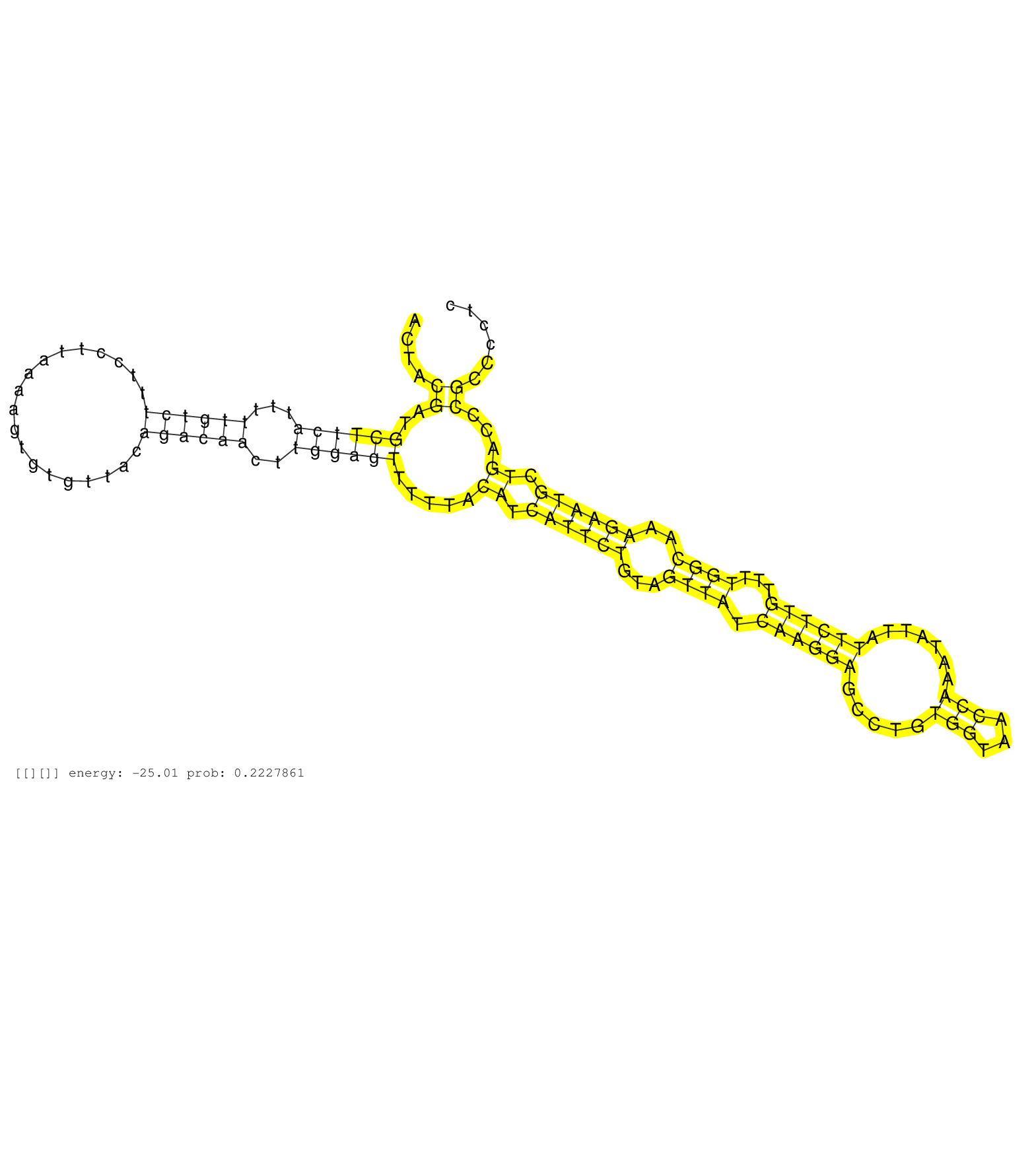
| ATCCGTGCAGCCTAAATACTGTACGCAGAGATCGGTCCTATAAACTCTGAACTACGATGCTTCATTTTTGTCTTTCCTTAAAAGTGTGTTACAGACAACTTGGAGTTTTTACATCATTCTGTAGTTATCAAGGAGCCTGTGGTAACCAAATATTATTCTTGTTTTGGCAAAGAATGCTGACCCGCCCCTCTGCTCCACAGTGCACTCTTTCTCCTGGTCATTGGAACAGCCTACTTGGAAGCTCAAGGGA ......................................................((..((((((...((((((...................))))))..)))))).....((.((((((...((((.((((((.....(((...))).......))))))...))))..)))))).))...)).................................................................. ..................................................51.........................................................................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................TTTTTACATCATTCTGTAGTTATCAAGGAGCCTGTGGTAACCAAATATTATT............................................................................................. | 52 | 1 | 28.00 | 28.00 | 28.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TTGGAACAGCCTACTTGGAAGCTCAA.... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - |
| ......................................................................................................................................................................................................................TGGTCATTGGAACAGCCTACTTGGAAGCTC...... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................................................................................................TGGTCATTGGAACAGCCTACTTGGAAcc........ | 28 | cc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGGTCATTGGAACAGCCTACTTGGAAGC........ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GTCATTGGAACAGCCTACTTGGAAGC........ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCCTGGTCATTGGAACAGCCTACTTG............. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TTTCTCCTGGTCATTGGAACAGCCT.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - |
| ATCCGTGCAGCCTAAATACTGTACGCAGAGATCGGTCCTATAAACTCTGAACTACGATGCTTCATTTTTGTCTTTCCTTAAAAGTGTGTTACAGACAACTTGGAGTTTTTACATCATTCTGTAGTTATCAAGGAGCCTGTGGTAACCAAATATTATTCTTGTTTTGGCAAAGAATGCTGACCCGCCCCTCTGCTCCACAGTGCACTCTTTCTCCTGGTCATTGGAACAGCCTACTTGGAAGCTCAAGGGA ......................................................((..((((((...((((((...................))))))..)))))).....((.((((((...((((.((((((.....(((...))).......))))))...))))..)))))).))...)).................................................................. ..................................................51.........................................................................................................................................190.......................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................CTACGATGCTTCActct.......................................................................................................................................................................................... | 17 | ctct | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - |
| ........................................................ATGCTTCATTTTTGTCTTT............................................................................................................................................................................... | 19 | 3 | 1.33 | 1.33 | - | - | - | - | 1.00 | - | - | - | - | 0.33 |
| ......................................................................TTCCTTAAAAGTGTGctg.................................................................................................................................................................. | 18 | ctg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................ATGCTTCATTTTTGTCTT................................................................................................................................................................................ | 18 | 4 | 0.25 | 0.25 | - | - | - | - | 0.25 | - | - | - | - | - |