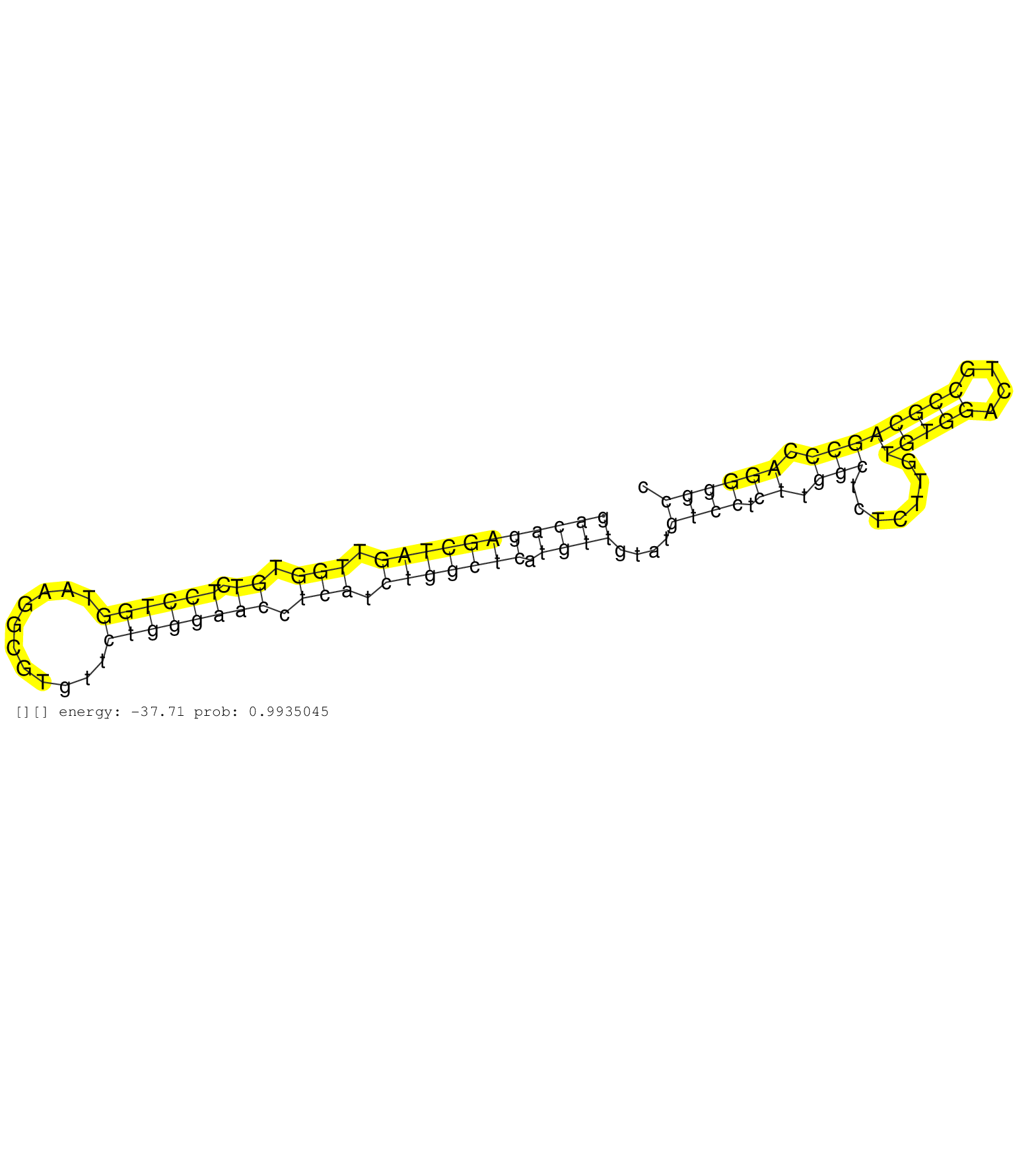

| Gene: Cnnm3 | ID: uc007aqi.1_intron_2_0_chr1_36577073_f | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(18) TESTES |
| AAAAATCTCGCCTCAGCTGCTCCTGGCCACCCAGCGCTTCCTTTCCCGAGGTGAGGGTGAAGGGTGGTCCTTGCCCCTGGTCTCACCACCCAGAGGGTCAAGAAACCACCCAGGCCAGACACCAGGCTCCTAGCGAGCTGGACAGAGCTAGTTGGTGTCTCCTGGTAAGGCGTGTTCTGGGAACCTCATCTGGCTCATGTTGTATGTCCTCTTGGCTCTCTTGTGTGGACTGCCGCAGCCCAGGGGCCACTGGGAGAGCCCAGGAGGCAGCAGGAGGCAGGTGTGATGACAGGCCATTTGGGCCCCAGTGGACACTGTCCCTTTCTCCCACCGTGGGGCAGAGGTGGATGTGTTCAGCCCATTGCGAGTCTCTGAGAAAGTCTTGCTGCAC ............................................................................................................................................(((((((((((.(((.((.((((((...........)))))))).))).))))))).))))....((((.((.(((.......(((((....)))))))).))))))................................................................................................................................................ ............................................................................................................................................141........................................................................................................248............................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................AGCTAGTTGGTGTCTCCTGGTAAGGCGT.......................................................................................................................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 1.00 | - | - | 2.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................GGTGTCTCCTGGTAAGGCGTGTTCTG.................................................................................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................AGAGGTGGATGTGTTCAGC................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GACAGAGCTAGTTGGTGTCTCCTGGT................................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TCTTGTGTGGACTGCCGCAGCCCAGG................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................GTTGGTGTCTCCTGGTAAGGCGTGTT....................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ....ATCTCGCCTCAGCTGCTCCTGGCCACC........................................................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................................................TGGGAGAGCCCAGGAGGCAGCAGGAGGC................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TGTTGTATGTCCTCTTGGCTCTCTTGTGT..................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................TCTCCTGGTAAGGCGTGTTCTGGGAA................................................................................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .AAAATCTCGCCTCAGCTGCTCCTGGCC........................................................................................................................................................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................GTGGATGTGTTCAGCCCATTGCGAGTC..................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TGGACAGAGCTAGTTGGTGTCTCCTG................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................TAGTTGGTGTCTCCTGGTAAGGCGTG......................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AGCGAGCTGGACAGAGCTAGTTGGT........................................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................................................................................................................................................GGGCAGAGGTGGATGgac...................................... | 18 | gac | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GCTGGACAGAGCTAGTTGGTGTCTCC..................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TGTTCTGGGAACCTCATCTGGCTCATG................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGCTAGTTGGTGTCTCCTGGTAA............................................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................TAGCGAGCTGGACAGAGCTAGTTGGTGT......................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................GGTGGATGTGTTCAGCCC............................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TAGCGAGCTGGACAGAGCTAGTTGGTG.......................................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................AGGTGTGATGACAGGCCATTTGGGCCCCA.................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..AAATCTCGCCTCAGCTGCTCCTG.............................................................................................................................................................................................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGTGTGGACTGCCGCAGCCCAGGGGtca.............................................................................................................................................. | 28 | tca | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................GAGGTGGATGTGTTCAGCCCATTGC.......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................................................................................GACAGAGCTAGTTGGTGTCTCCTGGTA................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................................................................................................................CCATTGCGAGTCTCTGAGAAAGTCTT....... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| AAAAATCTCGCCTCAGCTGCTCCTGGCCACCCAGCGCTTCCTTTCCCGAGGTGAGGGTGAAGGGTGGTCCTTGCCCCTGGTCTCACCACCCAGAGGGTCAAGAAACCACCCAGGCCAGACACCAGGCTCCTAGCGAGCTGGACAGAGCTAGTTGGTGTCTCCTGGTAAGGCGTGTTCTGGGAACCTCATCTGGCTCATGTTGTATGTCCTCTTGGCTCTCTTGTGTGGACTGCCGCAGCCCAGGGGCCACTGGGAGAGCCCAGGAGGCAGCAGGAGGCAGGTGTGATGACAGGCCATTTGGGCCCCAGTGGACACTGTCCCTTTCTCCCACCGTGGGGCAGAGGTGGATGTGTTCAGCCCATTGCGAGTCTCTGAGAAAGTCTTGCTGCAC ............................................................................................................................................(((((((((((.(((.((.((((((...........)))))))).))).))))))).))))....((((.((.(((.......(((((....)))))))).))))))................................................................................................................................................ ............................................................................................................................................141........................................................................................................248............................................................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................................................................................................................CCCACCGTGGGGCcg.................................................... | 15 | cg | 4.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| ..........................................................................................................................................................................................................................................................................................................................................................TGTTCAGCCCATaaaa............................. | 16 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................................................................................................................................................................................................CCCACCGTGGGGCAcgcc................................................... | 18 | cgcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |