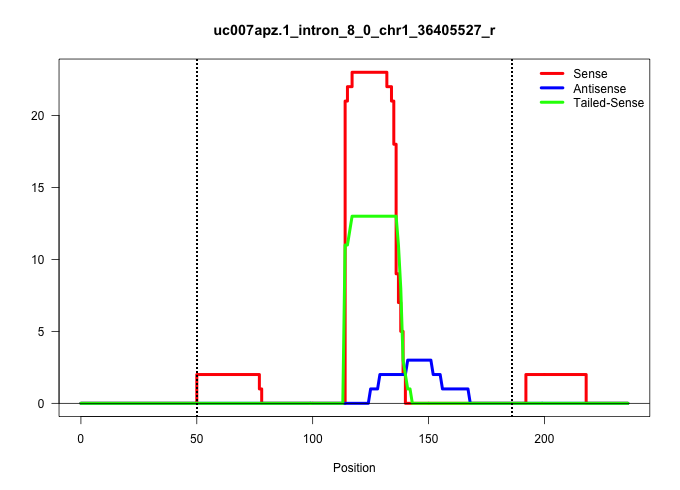
| Gene: 4632411B12Rik | ID: uc007apz.1_intron_8_0_chr1_36405527_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
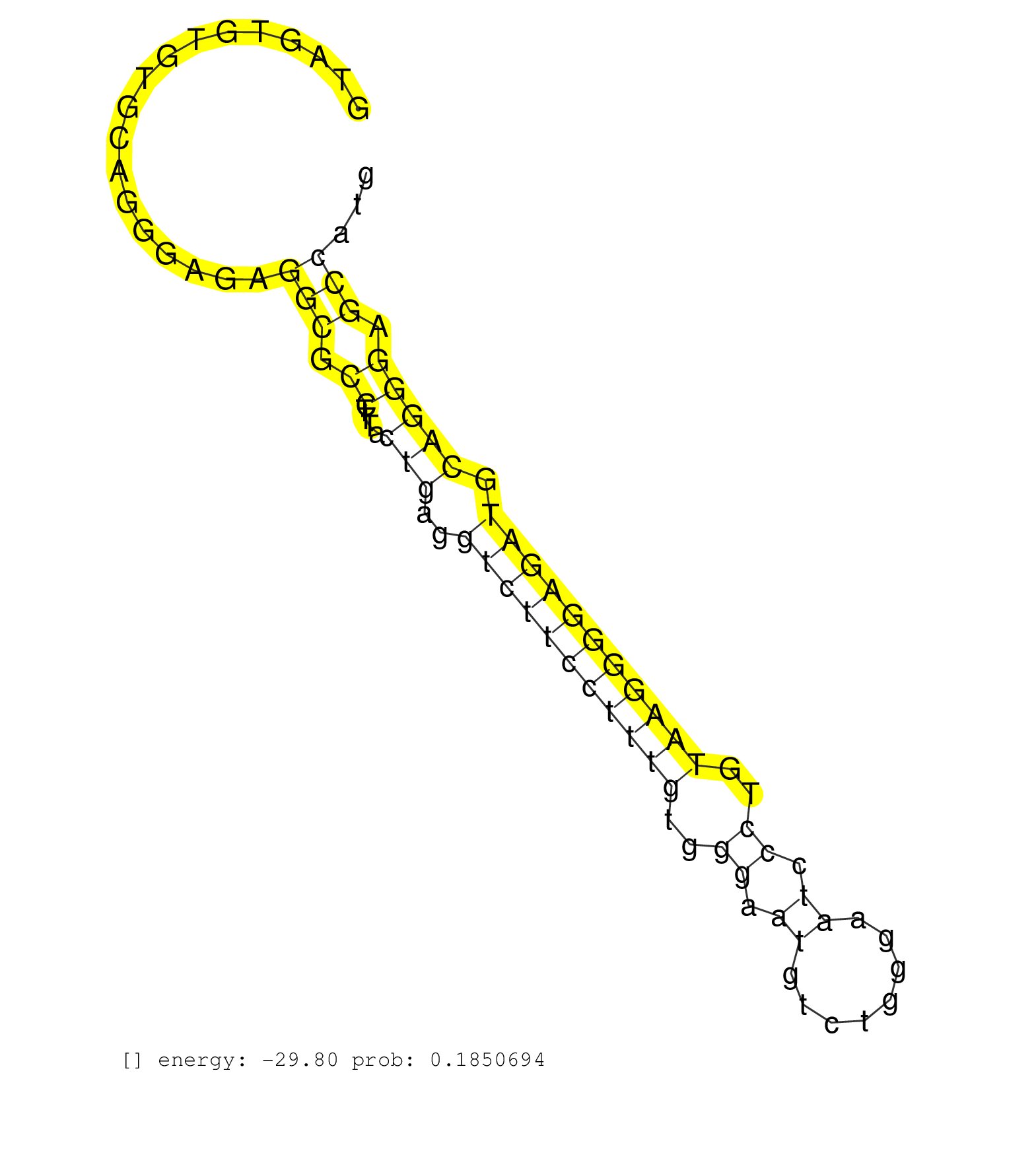
| GTCCTGCTTCCCCAGCTGCCAGGCTCCCCACATCCCCCTCAGGCTCAGAGGTAGTGTGTGCAGGGAGAGGCGCCTTTACTGAGGTCTTCCTTTGTGGGAATGTCTGGGAATCCCTGTAAGGGGAGATGCAGGGAGCCATGCCGAATGATGCTTGGATTCAAGTCTCTTTGTACCTTCTCTACCCAGGATCTCTCCAGTGTGTCCAGCAGCCCCACCTCTAGCCCTAAGACCAAAGT ....................................................................(((.((....(((..(((((((((((..((.((........)).))..))))))))))).))))).)))................................................................................................... ..................................................51.......................................................................................140.............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGC.................................................................................................... | 22 | 1 | 9.00 | 9.00 | 2.00 | 3.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCAT................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAG..................................................................................................... | 21 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCC................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| ................................................................................................................................................................................................TCCAGTGTGTCCAGCAGCCCCACCTC.................. | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCA.................................................................................................. | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAttt................................................................................................... | 23 | ttt | 2.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGA...................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCgt................................................................................................. | 25 | gt | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCt.................................................................................................. | 24 | t | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCAgtt............................................................................................... | 27 | gtt | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCtt................................................................................................. | 25 | tt | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCtt.................................................................................................. | 24 | tt | 1.00 | 9.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGG........................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................GTAAGGGGAGATGCAGGGAGCCATG................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGtaa.................................................................................................. | 24 | taa | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAAGGGGAGATGCAGGGAGCCtt................................................................................................. | 23 | tt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................AAGGGGAGATGCAGGGAGCCATG................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................AAGGGGAGATGCAGGGAGCCATttta............................................................................................. | 26 | ttta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAGTGTGTGCAGGGAGAGGCGCCTTT............................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCAaa................................................................................................ | 26 | aa | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGTGTGTGCAGGGAGAGGCGCCTTTA.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCaat................................................................................................. | 25 | aat | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................TGTAAGGGGAGATGCAGGGAGCCta................................................................................................. | 25 | ta | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| GTCCTGCTTCCCCAGCTGCCAGGCTCCCCACATCCCCCTCAGGCTCAGAGGTAGTGTGTGCAGGGAGAGGCGCCTTTACTGAGGTCTTCCTTTGTGGGAATGTCTGGGAATCCCTGTAAGGGGAGATGCAGGGAGCCATGCCGAATGATGCTTGGATTCAAGTCTCTTTGTACCTTCTCTACCCAGGATCTCTCCAGTGTGTCCAGCAGCCCCACCTCTAGCCCTAAGACCAAAGT ....................................................................(((.((....(((..(((((((((((..((.((........)).))..))))))))))).))))).)))................................................................................................... ..................................................51.......................................................................................140.............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014237(GSM319961) 10 dpp MILI-KO total. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................CGAATGATGCTTGGATTCAAGTCTCTT.................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................AGGGAGCCATGCCGAATGATGCTTGGA................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................ATGCAGGGAGCCATGCCGAATGATGCT.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................................................................GTGTCCAGCAGCCCC....................... | 15 | 19 | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 |