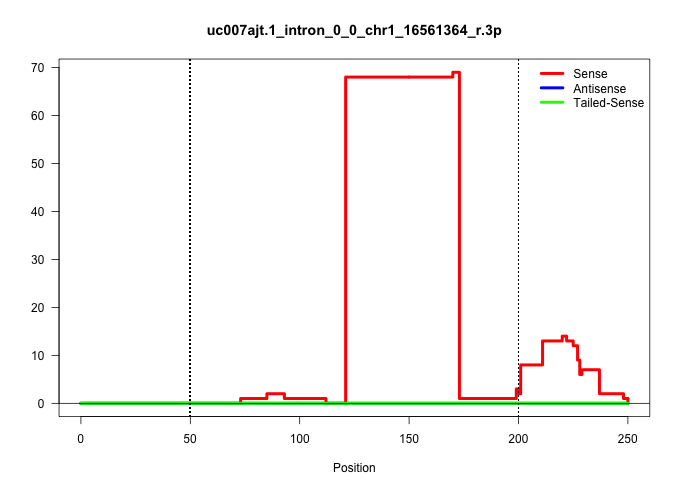
| Gene: Ube2w | ID: uc007ajt.1_intron_0_0_chr1_16561364_r.3p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(12) TESTES |
| AGCTCTCATTTCCTTGAAAGAGTAAAGGTTGGTTTGCTTATATTATATAACTGAACACAAATTTTCATCTGTAAGACTGAGAACGGCAAGTCAGGCTGGCCACGTTCTTGCTACTTGGCATTATTTCTCCATGTGATGAGCTTGTAGATGAACTTACACGTGCTTCCGTTTCTAATTCTCTTTTTCTCCTTTCCTTAAAGATGACACTTGTTGATGCCGCTGCCCTCCTCCTGTAGCAGAAGATAGTCCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................TATTTCTCCATGTGATGAGCTTGTAGATGAACTTACACGTGCTTCCGTTTCT............................................................................. | 52 | 1 | 68.00 | 68.00 | 68.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TGATGCCGCTGCCCTCCTCCTGTAGC............. | 26 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGACACTTGTTGATGCCGCTGCCCTC....................... | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................................................................................TGACACTTGTTGATGCCGCTGCCCTCC...................... | 27 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................................................TCTAATTCTCTTTTTCTCCTTTCCTTAAAG.................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................GATGACACTTGTTGATGCCGCTGCCC......................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................................................................................................................................................................................................CCTGTAGCAGAAGATAGTCCT | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGCCCTCCTCCTGTAGCAGAAGATAGTC.. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................GCAAGTCAGGCTGGCCACGTTCTTGCT.......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GATGACACTTGTTGATGCCGCTG............................ | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................AGACTGAGAACGGCAAGTCA............................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| AGCTCTCATTTCCTTGAAAGAGTAAAGGTTGGTTTGCTTATATTATATAACTGAACACAAATTTTCATCTGTAAGACTGAGAACGGCAAGTCAGGCTGGCCACGTTCTTGCTACTTGGCATTATTTCTCCATGTGATGAGCTTGTAGATGAACTTACACGTGCTTCCGTTTCTAATTCTCTTTTTCTCCTTTCCTTAAAGATGACACTTGTTGATGCCGCTGCCCTCCTCCTGTAGCAGAAGATAGTCCT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................TCTCCTTTCCTTAAAcca................................................... | 18 | cca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |