| (1) AGO2.ip | (3) B-CELL | (7) BRAIN | (1) CELL-LINE | (1) EMBRYO | (1) HEART | (1) KIDNEY | (5) LIVER | (4) LYMPH | (3) OTHER | (2) OTHER.mut | (1) PIWI.ip | (10) SPLEEN | (7) TESTES | (1) THYMUS | (1) UTERUS |
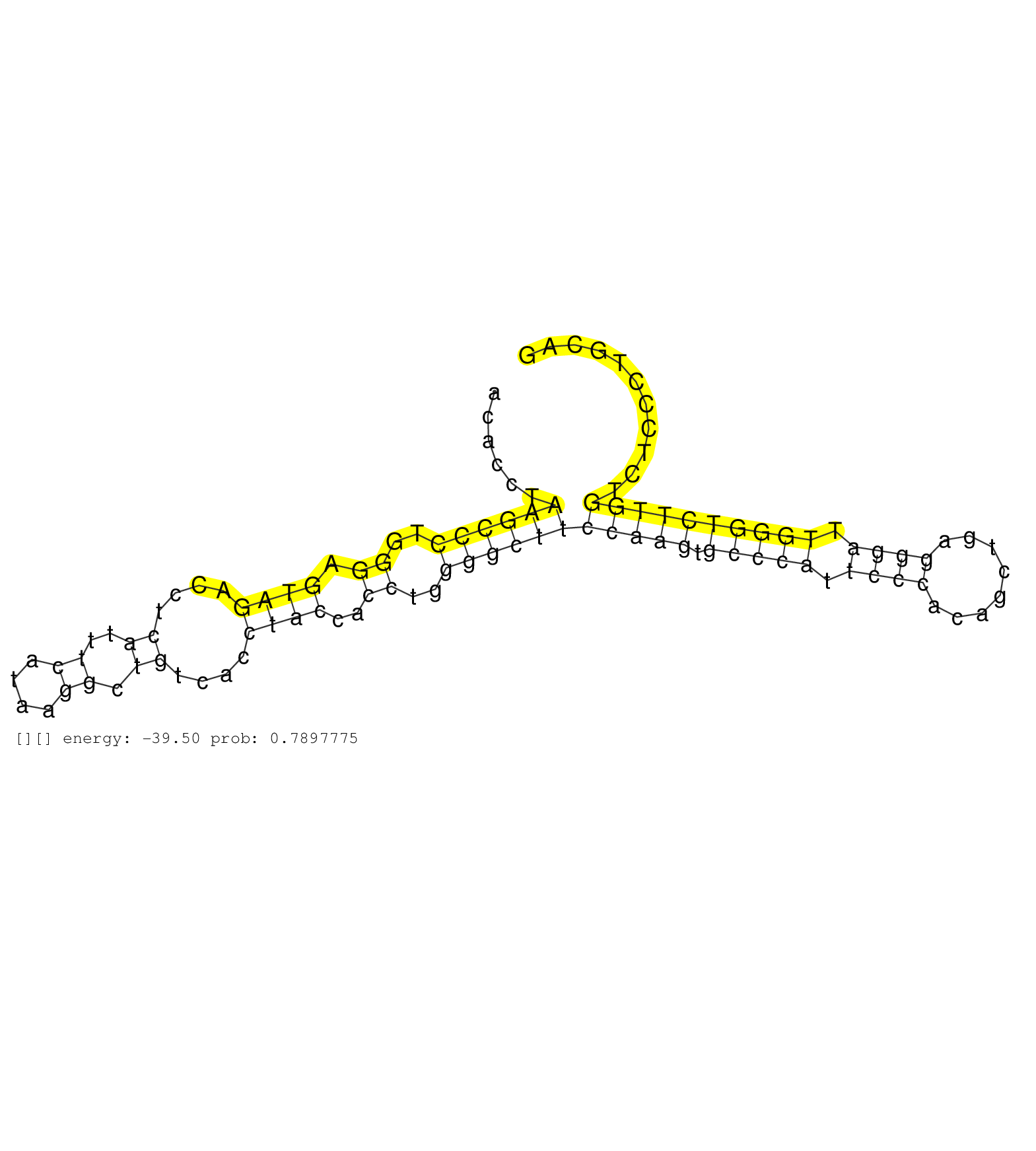
| AGCTGAGACTGGAAGTAGAGTCTTTAATCTCCCAGAAATACCTCATTTAGCCCCTGAGTCATTGGTGAACTCAAGACTCAGGCACATTGTACACCTAAGCCCTGGGAGTAGACCTCATTTCATAAGGCTGTCACCTACCACCTGGGGCTTCCAAGTGCCCATTCCCACAGCTGAGGGATTGGGTCTTGGTCTCCCTGCAGTTTGCATGACAACCAGCTCAAGGACCCAGAGGTGCTGAGTCTTGTGGAGC ................................................................................................((((((..((.((((....((..((....)).))....))))..))..))))))(((((.(((((.((((........)))).))))))))))............................................................. ..........................................................................................91...........................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR059769(GSM562831) Treg_Drosha. (spleen) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | mjLiverWT3() Liver Data. (liver) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR059768(GSM562830) Treg_control. (spleen) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR073954(GSM629280) total RNA. (blood) | mjTestesWT1() Testes Data. (testes) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM475281(GSM475281) total RNA. (testes) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR553582(SRX182788) source: Brain. (Brain) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR037902(GSM510438) testes_rep3. (testes) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAGT................................................. | 23 | 1 | 14.00 | 14.00 | 9.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAG.................................................. | 22 | 1 | 10.00 | 10.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCAGT................................................. | 24 | 1 | 5.00 | 5.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCAT.................................................. | 23 | 1 | 5.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAGA................................................. | 23 | 1 | 4.00 | 10.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAGTT................................................ | 24 | 1 | 4.00 | 4.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCT...................................................... | 19 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGGGTCTTGGTCTCCCTGCAGT................................................. | 22 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAT.................................................. | 22 | 1 | 3.00 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCAG.................................................. | 23 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCAGA................................................. | 24 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......ACTGGAAGTAGAGTCTATAG............................................................................................................................................................................................................................... | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCT...................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCA................................................... | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......ACTGGAAGTAGAGTCTTTGTG.............................................................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTTT.................................................... | 21 | 1 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGGGTCTTGGTCTCCCTGCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACTGGAAGTAGAGTCTGATG............................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAAGT................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TAAGCCCTGGGAGTAGAC......................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGT.................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGTT................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AAGGACCCAGAGGTGTTT............. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CATGACAACCAGCTCAAGGACCCAGA.................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACCTGGGGCTTCCAAGTGCC........................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACTGGAAGTAGAGTCGAAA................................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......ACTGGAAGTAGAGTCGTG................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..CTGAGACTGGAAGTATTCT..................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAGTAT............................................... | 25 | 1 | 1.00 | 14.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAATT................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACTGGAAGTAGAGTCTAAAG............................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTG..................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCAGTT................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CCCTGGGAGTAGACCGCA..................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................GCATGACAACCAGCTCAAGGACCCA...................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAGATT............................................... | 25 | 1 | 1.00 | 10.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACTGGAAGTAGAGTCAAA................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCAGTAT............................................... | 26 | 1 | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................GATTGGGTCTTGGTCTCCCTGT.................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCATAAT............................................... | 26 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......ACTGGAAGTAGAGTCTGAAG............................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTACAG.................................................. | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................ATTGGGTCTTGGTCTCCCTGCAGTTT............................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAGAAT............................................... | 25 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAAAA................................................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAA.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......ACTGGAAGTAGAGTCTGAA................................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................ACAACCAGCTCAAGGACCCA...................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTGGGTCTTGGTCTCCCTGCAGTAA............................................... | 25 | 1 | 1.00 | 14.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TAGCCCCTGAGTCATTGGTGA...................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................GTGAACTCAAGACTC........................................................................................................................................................................... | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCTGAGACTGGAAGTAGAGTCTTTAATCTCCCAGAAATACCTCATTTAGCCCCTGAGTCATTGGTGAACTCAAGACTCAGGCACATTGTACACCTAAGCCCTGGGAGTAGACCTCATTTCATAAGGCTGTCACCTACCACCTGGGGCTTCCAAGTGCCCATTCCCACAGCTGAGGGATTGGGTCTTGGTCTCCCTGCAGTTTGCATGACAACCAGCTCAAGGACCCAGAGGTGCTGAGTCTTGTGGAGC ................................................................................................((((((..((.((((....((..((....)).))....))))..))..))))))(((((.(((((.((((........)))).))))))))))............................................................. ..........................................................................................91...........................................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR059769(GSM562831) Treg_Drosha. (spleen) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | mjLiverWT3() Liver Data. (liver) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR059768(GSM562830) Treg_control. (spleen) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR073954(GSM629280) total RNA. (blood) | mjTestesWT1() Testes Data. (testes) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM475281(GSM475281) total RNA. (testes) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR553582(SRX182788) source: Brain. (Brain) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR037902(GSM510438) testes_rep3. (testes) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................ACAACCAGCTCAAGGGCC........................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................TCTCCCTGCAGTTTGATA........................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CCTGGGGCTTCCAAGGCAC........................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .......................................................................................................................................................................................................................GCTCAAGGACCCAGAAAA................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| .........................................................................................................................................CCACCTGGGGCTTCCAACA.............................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |