| (2) AGO2.ip | (20) BRAIN | (8) CELL-LINE | (2) DCR.mut | (1) DGCR8.mut | (1) EMBRYO | (1) ESC | (2) OTHER | (2) OTHER.mut | (1) SPLEEN | (3) TESTES |
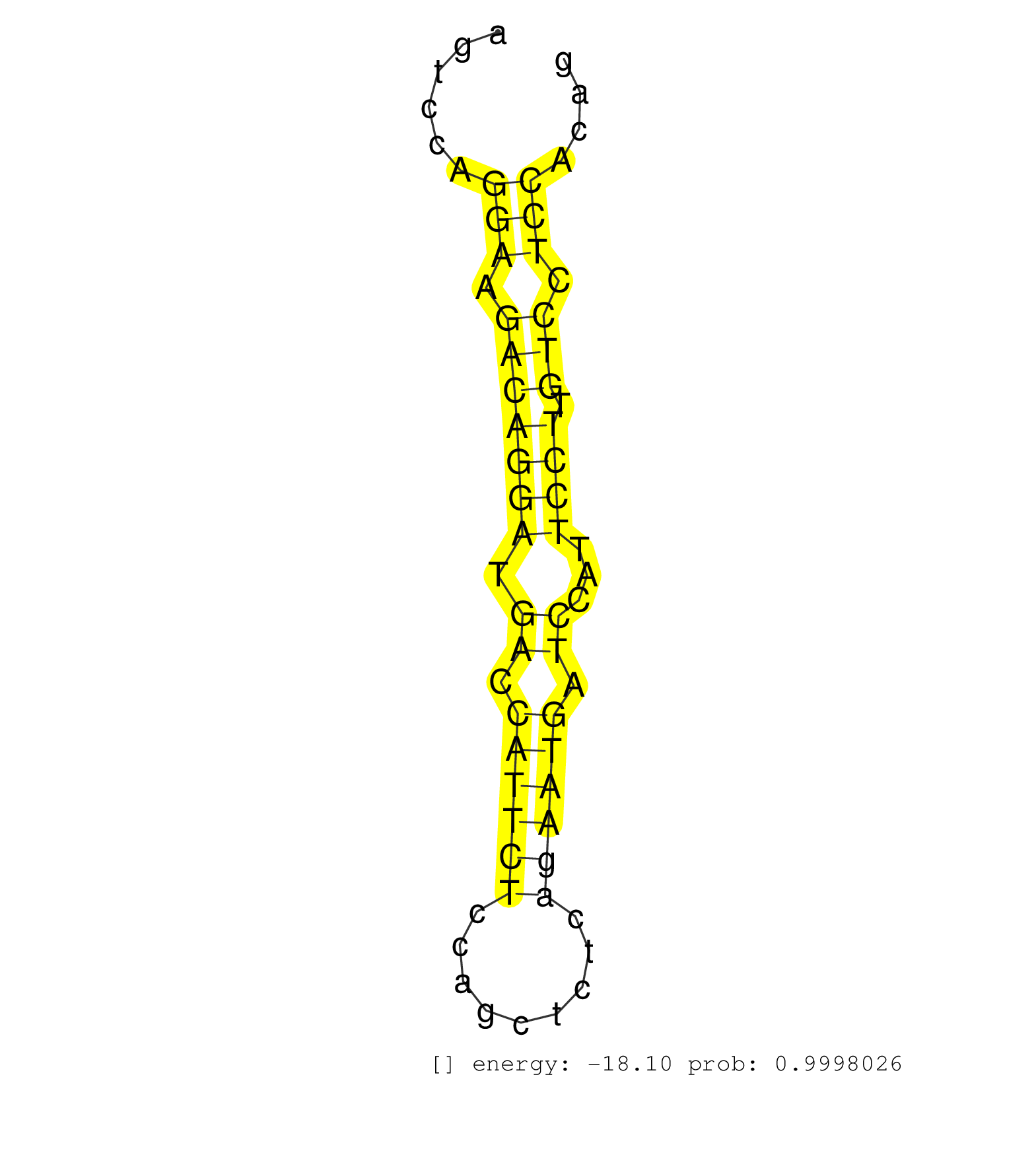
| AGAGTTATACTGCAGATAGAGGTTGCTGGCAGCATCTGGGGTGGAGTCCTGGTCAAGGCTCCACATTACATCAATCACAGCTCAGTGCCCACTACAGAGACCCATCAGCCTCAGTGCCAGAAATGTTGAGTTTAAGAGTCCAGGAAGACAGGATGACCATTCTCCAGCTCTCAGAATGATCCATTCCTTGTCCTCCACAGGGAACATGAGTTGGATCTCATGTCGGGGCATCTCTGGAGGCATTGAGGTC ..............................................................................................................................................(((.(((((((.((.((((((.........)))))).))...)))).))).)))...................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR206942(GSM723283) other. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR206941(GSM723282) other. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR073954(GSM629280) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR206940(GSM723281) other. (brain) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR037906(GSM510442) brain_rep3. (brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................AGGAAGACAGGATGACCATTCT....................................................................................... | 22 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTCCA..................................................... | 23 | 1 | 4.00 | 4.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTCC...................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................CAGGAAGACAGGATGACCATTC........................................................................................ | 22 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................ATGATCCATTCCTTGTCCTCCAC.................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................ATGATCCATTCCTTGTCCT........................................................ | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCC......................................................... | 19 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................ATGATCCATTCCTTGTCCTCCACAGT................................................. | 26 | 1 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AACATGAGTTGGATCCTGT............................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTCCACAGT................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTC....................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTCCAT.................................................... | 24 | 1 | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCAGGAAGACAGGATGACCATT......................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................TCTGGGGTGGAGTCCAC....................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................ATGATCCATTCCTTGTCCTCCACAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTCCAC.................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................ATGATCCATTCCTTGTCCTC....................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTCCACAG.................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................CATGAGTTGGATCTCATGTCGGGGC..................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................ATGATCCATTCCTTGTCCTCC...................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGGAAGACAGGATGACCATTC........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CAGGAAGACAGGATGACCATT......................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCAGGAAGACAGGATGCAGA........................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................AATGATCCATTCCTTGTCCTCCACT................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CAGGAAGACAGGATGACCAT.......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CCAGGAAGACAGGATGACCAT.......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................AGTTGGATCTCATGTCGG........................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAGTTATACTGCAGATAGAGGTTGCTGGCAGCATCTGGGGTGGAGTCCTGGTCAAGGCTCCACATTACATCAATCACAGCTCAGTGCCCACTACAGAGACCCATCAGCCTCAGTGCCAGAAATGTTGAGTTTAAGAGTCCAGGAAGACAGGATGACCATTCTCCAGCTCTCAGAATGATCCATTCCTTGTCCTCCACAGGGAACATGAGTTGGATCTCATGTCGGGGCATCTCTGGAGGCATTGAGGTC ..............................................................................................................................................(((.(((((((.((.((((((.........)))))).))...)))).))).)))...................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR206942(GSM723283) other. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR206941(GSM723282) other. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR073954(GSM629280) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR037935(GSM510473) 293cand3. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR037936(GSM510474) 293cand1. (cell line) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR206940(GSM723281) other. (brain) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR037906(GSM510442) brain_rep3. (brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................GAGTTGGATCTCATGTTGAG....................... | 20 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................ATGAGTTGGATCTCATGTAGC........................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................ACAGAGACCCATCAGCGGT.......................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .................................................................................................................................................................CTCCAGCTCTCAGAACTA....................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................TACAGAGACCCATCAGAA............................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................GAGTTGGATCTCATGTTGA........................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................CACTACAGAGACCCAACA............................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................CCCACTACAGAGACCCGTA................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................CAATCACAGCTCAGTGCCACC.............................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................TGAGTTGGATCTCATGTGAGC....................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................GCAGCATCTGGGGTGGACG........................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................ACATCAATCACAGCTC....................................................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .........................................................................................................................................................TGACCATTCTCCAGC.................................................................................. | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 |