| (1) AGO2.ip | (2) B-CELL | (6) BRAIN | (1) CELL-LINE | (1) EMBRYO | (1) ESC | (1) LIVER | (6) OTHER | (1) OTHER.mut | (1) OVARY | (2) SPLEEN | (6) TESTES | (2) TOTAL-RNA |
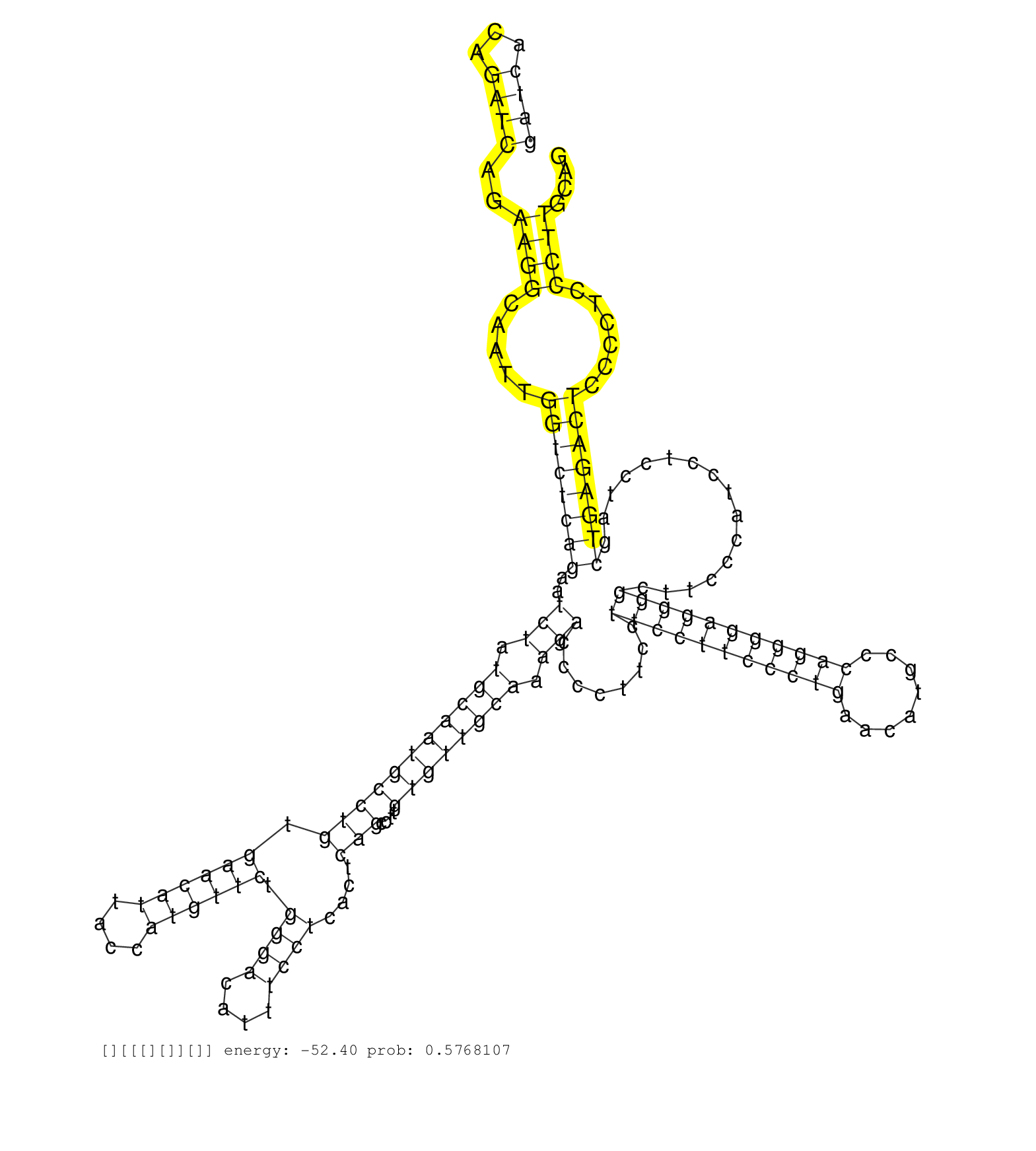
| TCTTCTTCGACGAAGACATCGGGGAAAATTCAGGAAAGATGGTGAGGAGGGTATGTGTAACTTGGGTCTCAGGAGGTGGTGTGTCTCCCTGTGAACATCGAAGCAGGCGCTCAGACACCAGCCATAGGGAAAGCTCAACCTGGGGCAGGGTAGCTTAGAAACGTGCAGATCACAGATCAGAAGGCAATTGGTCTCAGAATCTATGCAATGCCTGTGAACATTACCATGTTCTGGGACATTTCCTCACTCAGCCTTGTGTTGCAAAGACCCCTTCCTTCCTTCCCTGAACATGCCCAGGGGAGGGGCTTCCCATCCTCCTAGCTGAGACTCCCCTCCCTTGCAGTCCAGAAAGAGAAAGACTTGCAACTTTCTTCAGGAGCTGAAGAGCCCATA .......................................................................................................................................................................((((...))))..((((.....((((((((..(((.(((((((((((.((((((....)))))).((((....))))....)))....)))))))).)))........(((((((((((........)))))))))))................))))))))......))))...................................................... .......................................................................................................................................................................168............................................................................................................................................................................343................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | mjTestesWT1() Testes Data. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | mjTestesWT3() Testes Data. (testes) | SRR069834(GSM304914) Analysis of small RNAs in murine neutrophils cultured in vitro by Solexa/Illumina genome analyzer. (blood) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT2() Testes Data. (testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR037897(GSM510433) ovary_rep2. (ovary) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................................................................................................................TGAGACTCCCCTCCCTTGCAGT................................................. | 22 | 1 | 11.00 | 11.00 | 1.00 | 5.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................TGAGACTCCCCTCCCTTGCAGTAT............................................... | 24 | 1 | 5.00 | 11.00 | 1.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................TGAGACTCCCCTCCCTTGCAG.................................................. | 21 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................CAGATCAGAAGGCAACTGT.......................................................................................................................................................................................................... | 19 | 3.00 | 0.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................................................................................................................................CCAGAAAGAGAAAGATA................................ | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................................................................................................................................................AAGACTTGCAACTTTCTTC................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................TCAGGAGGTGGTGTGTGGAG................................................................................................................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TAGGGAAAGCTCAACGTG........................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................AGGAGGGTATGTGTAGGAT.......................................................................................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ............................................................................................................................................................................CAGATCAGAAGGCAACCG........................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........GACGAAGACATCGGGGAA............................................................................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................GTATGTGTAACTTGGGTCTCAGGAGGTGGTGTGTC.................................................................................................................................................................................................................................................................................................................... | 35 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CAGGGTAGCTTAGAAACGTGCA.................................................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................TGTGTTGCAAAGACCCCGTA....................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................................................CCCAGGGGAGGGGCTTCTTT................................................................................. | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................................................................................................................GAGACTCCCCTCCCTTGCAGT................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................CCCAGGGGAGGGGCTTCCCATCTAT............................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................TGAACATCGAAGCAGGCGCTCAGACACCAGC............................................................................................................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................AGGAAAGATGGTGAGAGTA....................................................................................................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................CAGATCAGAAGGCAAGTCT.......................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................................................CCCAGGGGAGGGGCTTCCCATCT.............................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................TGAGACTCCCCTCCCTTGCAGA................................................. | 22 | 1 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................CAGGAGGTGGTGTGTGCC.................................................................................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................AGGGAAAGCTCAACCTGGGGCAGGGTAGCTT............................................................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................TGAGACTCCCCTCCCTTGCCGT................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TTCAGGAAAGATGGTCTGC.......................................................................................................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................................................................................................................TGAGACTCCCCTCCCTTGCAGTA................................................ | 23 | 1 | 1.00 | 11.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................CCCAGGGGAGGGGCTTCCCATC............................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................TGAGACTCCCCTCCCTTGC.................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................TAGCTGAGACTCCCCTCCCTTG..................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................CATCGGGGAAAATTCAGGAAAGATGTC.............................................................................................................................................................................................................................................................................................................................................................. | 27 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................................TCCCTTGCAGTCCAG............................................. | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| TCTTCTTCGACGAAGACATCGGGGAAAATTCAGGAAAGATGGTGAGGAGGGTATGTGTAACTTGGGTCTCAGGAGGTGGTGTGTCTCCCTGTGAACATCGAAGCAGGCGCTCAGACACCAGCCATAGGGAAAGCTCAACCTGGGGCAGGGTAGCTTAGAAACGTGCAGATCACAGATCAGAAGGCAATTGGTCTCAGAATCTATGCAATGCCTGTGAACATTACCATGTTCTGGGACATTTCCTCACTCAGCCTTGTGTTGCAAAGACCCCTTCCTTCCTTCCCTGAACATGCCCAGGGGAGGGGCTTCCCATCCTCCTAGCTGAGACTCCCCTCCCTTGCAGTCCAGAAAGAGAAAGACTTGCAACTTTCTTCAGGAGCTGAAGAGCCCATA .......................................................................................................................................................................((((...))))..((((.....((((((((..(((.(((((((((((.((((((....)))))).((((....))))....)))....)))))))).)))........(((((((((((........)))))))))))................))))))))......))))...................................................... .......................................................................................................................................................................168............................................................................................................................................................................343................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | mjTestesWT1() Testes Data. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | mjTestesWT3() Testes Data. (testes) | SRR069834(GSM304914) Analysis of small RNAs in murine neutrophils cultured in vitro by Solexa/Illumina genome analyzer. (blood) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | mjTestesWT2() Testes Data. (testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (gland) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | SRR037897(GSM510433) ovary_rep2. (ovary) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................CAGATCACAGATCAGAAGGCAATTGGT......................................................................................................................................................................................................... | 27 | 1 | 7.00 | 7.00 | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................CAGACACCAGCCATAGGGAAAGCTCAAC.............................................................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TGTGTCTCCCTGTGATTTA....................................................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................................................................................................................................CCCTCCCTTGCAGTCCAGAAAGAGA...................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GCTTAGAAACGTGCAGATCACAGATCAGA.................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CGAAGCAGGCGCTC......................................................................................................................................................................................................................................................................................... | 14 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ..............................................................................................................................................................................................GTCTCAGAATCTATGC........................................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |