| (5) AGO2.ip | (1) B-CELL | (21) BRAIN | (3) CELL-LINE | (1) DGCR8.mut | (1) EMBRYO | (1) ESC | (1) HEART | (4) LIVER | (1) LYMPH | (2) OTHER | (3) OTHER.mut | (1) OVARY | (1) PIWI.mut | (1) SPLEEN | (7) TESTES | (1) TOTAL-RNA |
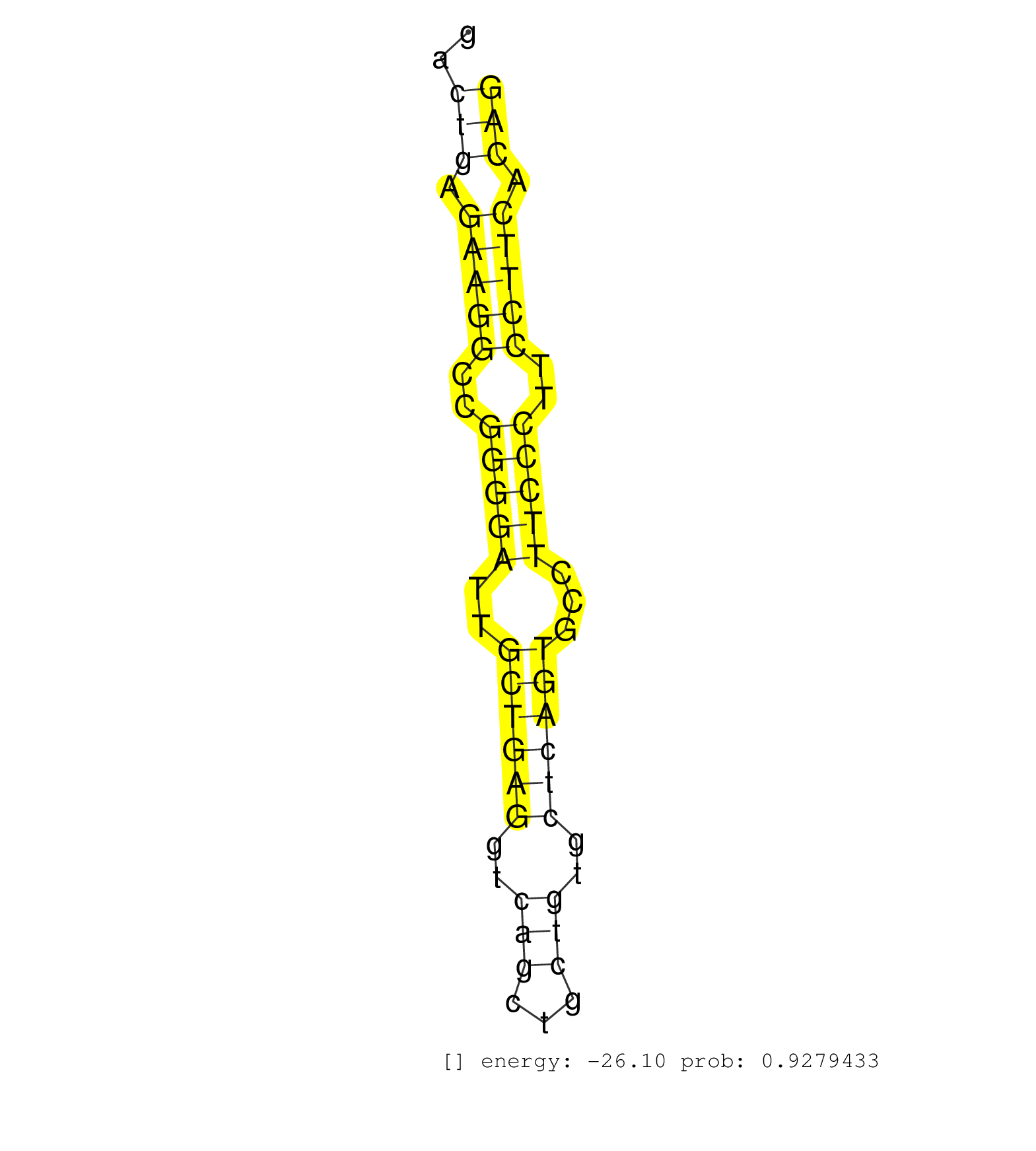
| AGGGATGACATCAGAGCTAAAGTCATAGGAAGGTCACTCTGCTAGAGAGCCAGCCAGAACAGGGTGTTCAGAAAGGTGGCAGAAGACAGTCCCTCGGAAGATGGGGGGGGGGGGGGGCGACACCCATAGCTCACAGGACTGAGAAGGCCGGGGATTGCTGAGGTCAGCTGCTGTGCTCAGTGCCTTCCCTTCCTTCACAGACACTTCATCATCATCGATGAGGAGAAGTTTCGGGAGATTTGGCTGATGA ..........................................................................................................................................(((.(((((..(((((..((((((..(((...)))..))))))...)))))..))))).))).................................................. ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR073954(GSM629280) total RNA. (blood) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR206942(GSM723283) other. (brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR553584(SRX182790) source: Heart. (Heart) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR065045(SRR065045) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR060845(GSM561991) total RNA. (brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR014234(GSM319958) Ovary total. (ovary) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGAG........................................................................................ | 21 | 1 | 13.00 | 13.00 | 4.00 | 2.00 | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGAAGGCCGGGGATTGCTGA......................................................................................... | 22 | 1 | 9.00 | 9.00 | 1.00 | 1.00 | - | - | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGCTGTGCTCAGTGCATT................................................................ | 18 | 8.00 | 0.00 | - | - | 5.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AGTGCCTTCCCTTCCTTCACAGT................................................. | 23 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGA......................................................................................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGAGA....................................................................................... | 22 | 1 | 2.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGAAGGCCGGGGATTG............................................................................................. | 18 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGAAGGCCGGGGATTGCT........................................................................................... | 20 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..........................................................................................................................................CTGAGAAGGCCGGGGATTGCTGA......................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGAGT....................................................................................... | 22 | 1 | 2.00 | 13.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGAAGGCCGGGGATTGCTG.......................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTG.......................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGGG........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TCAGTGCCTTCCCTTCCTTCACA................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................ATCGATGAGGAGAAGGAGG.................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................TGAGAAGGCCGGGGATTGTTGT......................................................................................... | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................CAGTGCCTTCCCTTCCTTCACA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGAAGGCCGGGGATTGCTGATA....................................................................................... | 24 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGAGAT...................................................................................... | 23 | 1 | 1.00 | 13.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GGGGGGGGGGGGGGCGAAAAG.............................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................TAGCTCACAGGACTGAGAAGGCCGGG.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GGATGACATCAGAGCAAC...................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ..............................................................................................................................TAGCTCACAGGACTGATG.......................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGAAG....................................................................................... | 22 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................TGAGAAGGCCGGGGATTGTTA.......................................................................................... | 21 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CTCACAGGACTGAGACGT....................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................TGAGGAGAAGTTTCGGGAGATTTGGA...... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CACAGGACTGAGAAGGCTA.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................TCATCATCATCGATGTCGC.......................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................TGCTAGAGAGCCAGCCAGAACAGGGCG........................................................................................................................................................................................ | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .............................................................................................................................................AGAAGGCCGGGGATTGCT........................................................................................... | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................TCATCGATGAGGAGAAGTTTCGGGAG............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGCTAGAGAGCCAGCCAGAACAGGGTGT....................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TCATCATCGATGAGGAGAAGTTTCG................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GAGAAGGCCGGGGATTGCTG.......................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................ATCGATGAGGAGAAGCC.................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................AGGCCGGGGATTGCTGAGA....................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................CGGAAGATGGGGGGGTA........................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................AGAAGGCCGGGGATTGCTGTG........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CGGAAGATGGGGGGGGGG.......................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ....................................................................................................................................ACAGGACTGAGAAGGC...................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGGATGACATCAGAGCTAAAGTCATAGGAAGGTCACTCTGCTAGAGAGCCAGCCAGAACAGGGTGTTCAGAAAGGTGGCAGAAGACAGTCCCTCGGAAGATGGGGGGGGGGGGGGGCGACACCCATAGCTCACAGGACTGAGAAGGCCGGGGATTGCTGAGGTCAGCTGCTGTGCTCAGTGCCTTCCCTTCCTTCACAGACACTTCATCATCATCGATGAGGAGAAGTTTCGGGAGATTTGGCTGATGA ..........................................................................................................................................(((.(((((..(((((..((((((..(((...)))..))))))...)))))..))))).))).................................................. ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR073954(GSM629280) total RNA. (blood) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR206942(GSM723283) other. (brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR553584(SRX182790) source: Heart. (Heart) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR065045(SRR065045) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR060845(GSM561991) total RNA. (brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR014234(GSM319958) Ovary total. (ovary) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................TTGCTGAGGTCAGCTAGC.............................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................GAGCCAGCCAGAACAGACA......................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................................TTCATCATCATCGATAAAA........................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................GGTCAGCTGCTGTGCGC........................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................CTGTGCTCAGTGCCTGGA.............................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................CAGACACTTCATCATCATCGATGAGGA.......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................CAGTGCCTTCCCTTCCTGT...................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................TGAGAAGGCCGGGGACC.............................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................GCCAGAACAGGGTGTGCCT................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |