| (1) AGO1.ip | (1) AGO2.ip | (2) AGO3.ip | (9) BRAIN | (1) DGCR8.mut | (1) EMBRYO | (1) ESC | (1) HEART | (1) LIVER | (3) OTHER | (4) SKIN | (1) TESTES | (1) UTERUS |
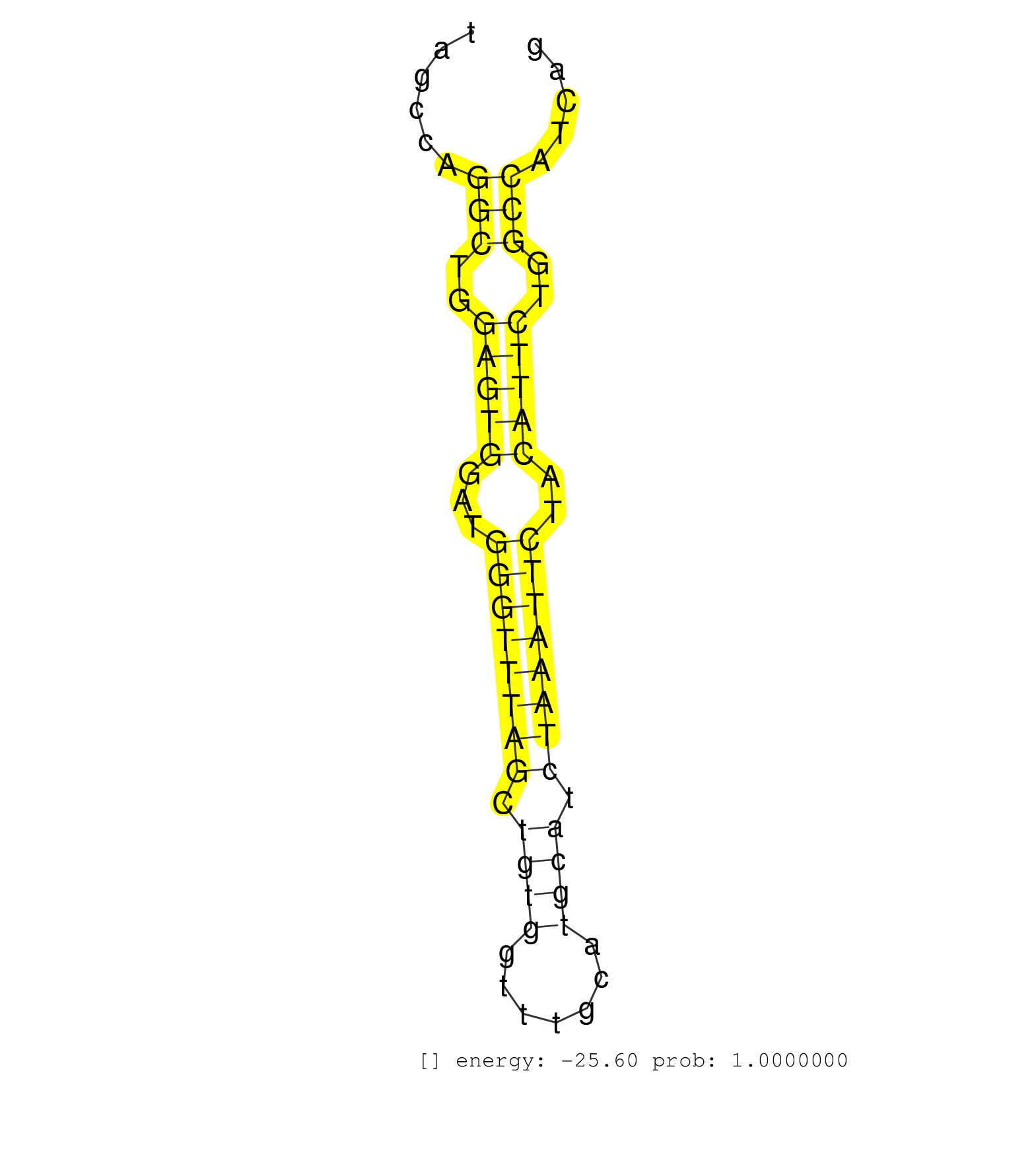
| GGTACAGAATGATGGTGATGGGTATCTGCTCTGTGCTAGACCAATGGGTCACAGTTAGAGCTGAGGTCTGGCTGGAGTAAAGGCAGAGGAGTGCTATGTTGTCATATCAGTATGCCCCCAGAGTCAGATCCTAGCCAGGCTGGAGTGGATGGGTTTAGCTGTGGTTTGCATGCATCTAAATTCTACATTCTGGCCATCAGGTGTGTTCCAGTGCGGTCCAGCTTCCGTTAATGCAATCAAAGCCGGTGAT .........................................................................................................................................(((..(((((...((((((((.((((.......)))).))))))))..)))))..)))....................................................... ...................................................................................................................................132.................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR073954(GSM629280) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR206942(GSM723283) other. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | GSM510444(GSM510444) brain_rep5. (brain) | mjLiverWT1() Liver Data. (liver) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR095855BC3(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................AGGCTGGAGTGGATGGGTTTAGC........................................................................................... | 23 | 1 | 15.00 | 15.00 | 4.00 | 3.00 | 4.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAAATTCTACATTCTGGCCATC.................................................... | 22 | 1 | 5.00 | 5.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ........................................................................................................................................AGGCTGGAGTGGATGGGTTTAG............................................................................................ | 22 | 1 | 5.00 | 5.00 | 2.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CAGGCTGGAGTGGATGGGTTTAGC........................................................................................... | 24 | 1 | 3.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CAGGCTGGAGTGGATGGGTTTCGC........................................................................................... | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AGGCTGGAGTGGATGGGTTTAGT........................................................................................... | 23 | 1 | 2.00 | 5.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AAATTCTACATTCTGACTA...................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................AGGCTGGAGTGGATGGGTTTAGCT.......................................................................................... | 24 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CAGGCTGGAGTGGATGGGTTT.............................................................................................. | 21 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CAGGCTGGAGTGGATGGGTTTCG............................................................................................ | 23 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAAATTCTACATTCTGGCCATCA................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CAGGCTGGAGTGGATGGGTTTAGCG.......................................................................................... | 25 | 1 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GGCAGAGGAGTGCTATCCTT..................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................CCAGGCTGGAGTGGATGGGTTTAGC........................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GGCAGAGGAGTGCTATACA...................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ...........................................................................................................................................CTGGAGTGGATGGGTTTAGC........................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GGCAGAGGAGTGCTACAC....................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................AGAGCTGAGGTCTGGCTGGCG............................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TAAATTCTACATTCTGGCC....................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................GTTAGAGCTGAGGTCTG.................................................................................................................................................................................... | 17 | 4 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CAGGCTGGAGTGGATGGGTTTAG............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GGCAGAGGAGTGCTATGCT...................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................AGGCTGGAGTGGATGGGT................................................................................................ | 18 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| GGTACAGAATGATGGTGATGGGTATCTGCTCTGTGCTAGACCAATGGGTCACAGTTAGAGCTGAGGTCTGGCTGGAGTAAAGGCAGAGGAGTGCTATGTTGTCATATCAGTATGCCCCCAGAGTCAGATCCTAGCCAGGCTGGAGTGGATGGGTTTAGCTGTGGTTTGCATGCATCTAAATTCTACATTCTGGCCATCAGGTGTGTTCCAGTGCGGTCCAGCTTCCGTTAATGCAATCAAAGCCGGTGAT .........................................................................................................................................(((..(((((...((((((((.((((.......)))).))))))))..)))))..)))....................................................... ...................................................................................................................................132.................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR073954(GSM629280) total RNA. (blood) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR206942(GSM723283) other. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | GSM510444(GSM510444) brain_rep5. (brain) | mjLiverWT1() Liver Data. (liver) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR095855BC3(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................ATCTGCTCTGTGCTAGACC................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................TCATATCAGTATGCCCCCAGAGTCAGA.......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....................................................GTTAGAGCTGAGGTCGT.................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................TGTTGTCATATCAGTA.......................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ...............................TGTGCTAGACCAATG............................................................................................................................................................................................................ | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |