| (1) AGO.mut | (5) AGO2.ip | (23) BRAIN | (3) CELL-LINE | (3) DCR.mut | (2) DGCR8.mut | (8) EMBRYO | (3) ESC | (2) FIBROBLAST | (3) LIVER | (1) OTHER | (2) OTHER.mut | (1) TESTES | (2) TOTAL-RNA |
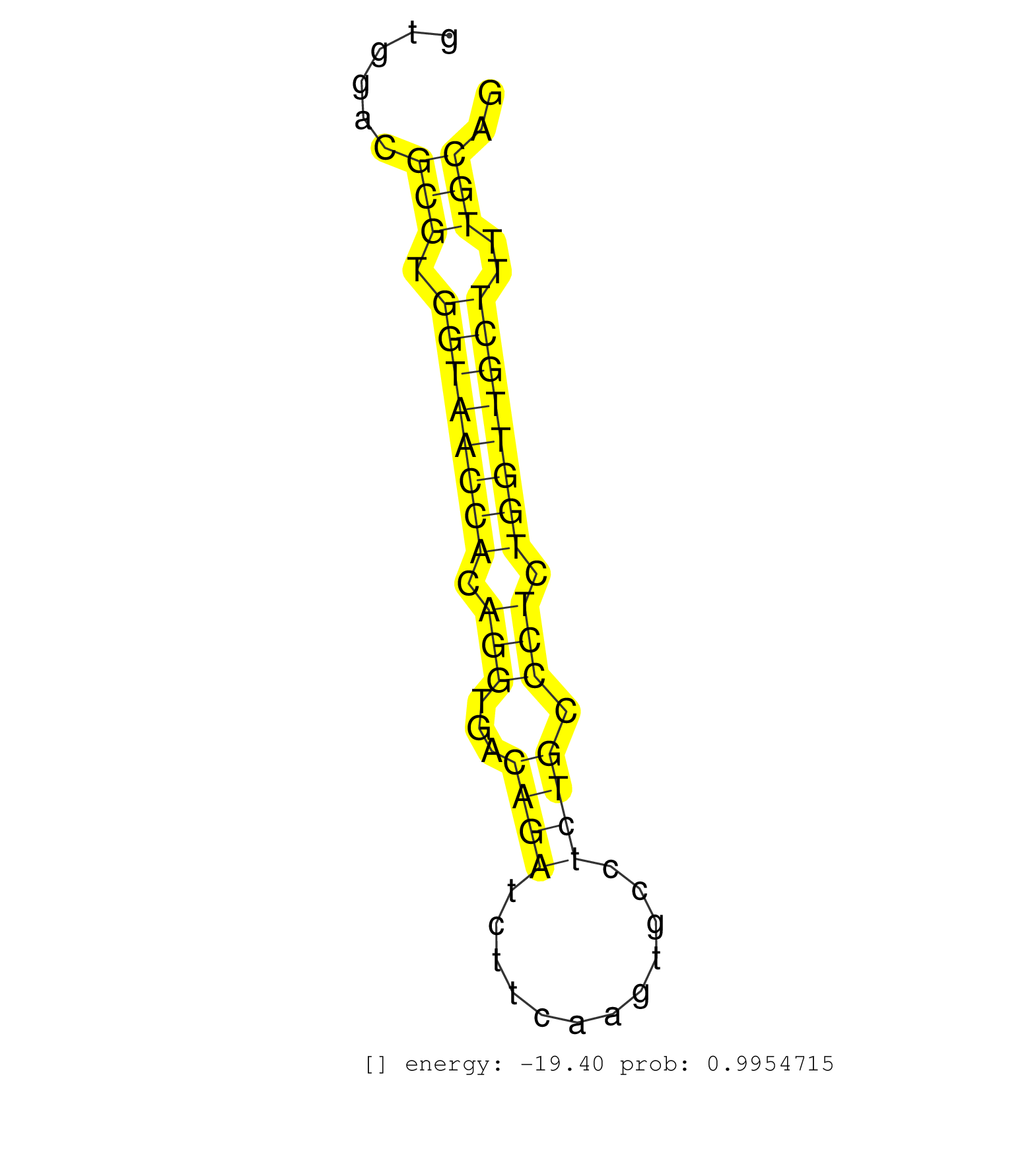
| CAGTGAGCCCTAGGCCTGGCCTAGAGGGAAGGTGGCTTCCTGTACGACATCTCTCCTCAGAGCTGGGCTACCTCCGTCTAGGGCAGGGAAGGGTAGCAGCTTTGGCATGACTGTCCTCAGGATGCTCTGGCTACTGTGGACGCGTGGTAACCACAGGTGACAGATCTTCAAGTGCCTCTGCCCTCTGGTTGCTTTTGCAGTTGAGAGCACATGCAGTGGACATCAATGGCAATCAAGTGGAGAACCCCAT .............................................................................................................................................(((.((((((((.(((...((((............)))).))).))))))))..))).................................................... .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR394084(GSM855970) "background strain: C57BL6/SV129cell type: KR. (dicer cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR206940(GSM723281) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | GSM361408(GSM361408) WholeCerebellum_P6_wt_rep1. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM361407(GSM361407) CGNP_P6_Ptc+-_Ink4c--_rep5. (brain) | GSM361430(GSM361430) WholeCerebellum_1month_Ptc+-_Ink4c--_rep2. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR060845(GSM561991) total RNA. (brain) | SRR206941(GSM723282) other. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | GSM361415(GSM361415) WholeCerebellum_P6_p53--_Ink4c--_rep4. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TGCCCTCTGGTTGCTTTTGCAGT................................................. | 23 | 1 | 21.00 | 21.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - |
| ..................................................................................................................................................................................TGCCCTCTGGTTGCTTTTGCAG.................................................. | 22 | 1 | 10.00 | 10.00 | - | - | 1.00 | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GAGAGCACATGCAGTGGAC............................. | 19 | 1 | 5.00 | 5.00 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................ATGCAGTGGACATCAATGG..................... | 19 | 1 | 5.00 | 5.00 | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CGCGTGGTAACCACAGGTGACAGA...................................................................................... | 24 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................AGTGGACATCAATGGCA................... | 17 | 2 | 3.50 | 3.50 | - | - | - | - | - | - | - | 3.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................ACGACATCTCTCCTCAGAGCT.......................................................................................................................................................................................... | 21 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ATGACTGTCCTCAGGAGATT............................................................................................................................ | 20 | 3.00 | 0.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................GGACATCAATGGCAATCAAGT............ | 21 | 1 | 3.00 | 3.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................AGTGGACATCAATGGCAATCAAGT............ | 24 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGCACATGCAGTGGACACC.......................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................GAGAGCACATGCAGTGGACAT........................... | 21 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................ATGGCAATCAAGTGGAGAACCCCA. | 24 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GAGAGCACATGCAGTGGACATC.......................... | 22 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GCGTGGTAACCACAGGTGACAGA...................................................................................... | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACGCGTGGTAACCACAGGTGAC......................................................................................... | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAGCACATGCAGTGGACATCAAT....................... | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................ACGACATCTCTCCTCAGAGCTGG........................................................................................................................................................................................ | 23 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TGGCAATCAAGTGGAGAACCCC.. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TTGAGAGCACATGCAGTGGACAT........................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACGCGTGGTAACCACAGGTGACAGA...................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GGACGCGTGGTAACCACAGGTGACAGATC.................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................TGCCCTCTGGTTGCTTTTGCAGTA................................................ | 24 | 1 | 1.00 | 21.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................ATGCTCTGGCTACTGTGGACGCGTGGTAACCACAGGTGACAGATCTTCAAGTGCCTCTGCCCTCTGGTTGCTTTTGCAG.................................................. | 79 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................GGATGCTCTGGCTACTGTGGACGCGT......................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GGCAATCAAGTGGAGAACCCCA. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GTGGACATCAATGGCAATCAAGTGGAGAA...... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GTGGACATCAATGGCAAT................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................GGCAATCAAGTGGAGAACCCC.. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGCCCTCTGGTTGCTTTTGCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GCGTGGTAACCACAGGTGACAG....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................AATGGCAATCAAGTGGAGAACCCC.. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGACATCAATGGCAATCAAG............. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GGGAAGGGTAGCAGCTTTGGCATGAC........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................................................................................................GAGAGCACATGCAGTGGACATCAA........................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CAATGGCAATCAAGTGGAGAACCCC.. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACGCGTGGTAACCACAGGTGACAG....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TGAGAGCACATGCAGTGGACATCAA........................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGACATCAATGGCAATCA............... | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAGTGAGCCCTAGGCCTGGCCTAGAGGGAAGGTGGCTTCCTGTACGACATCTCTCCTCAGAGCTGGGCTACCTCCGTCTAGGGCAGGGAAGGGTAGCAGCTTTGGCATGACTGTCCTCAGGATGCTCTGGCTACTGTGGACGCGTGGTAACCACAGGTGACAGATCTTCAAGTGCCTCTGCCCTCTGGTTGCTTTTGCAGTTGAGAGCACATGCAGTGGACATCAATGGCAATCAAGTGGAGAACCCCAT .............................................................................................................................................(((.((((((((.(((...((((............)))).))).))))))))..))).................................................... .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR394084(GSM855970) "background strain: C57BL6/SV129cell type: KR. (dicer cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR206940(GSM723281) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | GSM361408(GSM361408) WholeCerebellum_P6_wt_rep1. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | GSM361407(GSM361407) CGNP_P6_Ptc+-_Ink4c--_rep5. (brain) | GSM361430(GSM361430) WholeCerebellum_1month_Ptc+-_Ink4c--_rep2. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR060845(GSM561991) total RNA. (brain) | SRR206941(GSM723282) other. (brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | GSM361415(GSM361415) WholeCerebellum_P6_p53--_Ink4c--_rep4. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR685340(GSM1079784) "Small RNAs (15-50 nts in length) from immort. (dicer cell line) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR037906(GSM510442) brain_rep3. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR023849(GSM307158) NPCsmallrna_rep1. (cell line) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................TGCCCTCTGGTTGCTTTTGCAGT................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGCCCTCTGGTTGCTTTTGCAG.................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGCCCTCTGGTTGCTTTG...................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................TCCTCAGAGCTGGGCTTTT.................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CGCGTGGTAACCACAGGTGACAGAT..................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................AGTGCCTCTGCCCTCGCT.............................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................GATGCTCTGGCTACTTCC................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................GGCTTCCTGTACGACGGTA...................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................GTGCCTCTGCCCTCTGGTGGT.......................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |